Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID-19) diagnosis and treatment using microarray dataset and bioinformatics methods
- PMID: 33251083
- PMCID: PMC7679428
- DOI: 10.1007/s13205-020-02406-y
Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID-19) diagnosis and treatment using microarray dataset and bioinformatics methods
Abstract
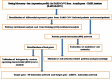
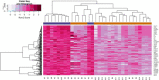
The goal of the present investigation is to identify the differentially expressed genes (DEGs) between SARS-CoV-2 infected and normal control samples to investigate the molecular mechanisms of infection with SARS-CoV-2. The microarray data of the dataset E-MTAB-8871 were retrieved from the ArrayExpress database. Pathway and Gene Ontology (GO) enrichment study, protein-protein interaction (PPI) network, modules, target gene-miRNA regulatory network, and target gene-TF regulatory network have been performed. Subsequently, the key genes were validated using an analysis of the receiver operating characteristic (ROC) curve. In SARS-CoV-2 infection, a total of 324 DEGs (76 up- and 248 down-regulated genes) were identified and enriched in a number of associated SARS-CoV-2 infection pathways and GO terms. Hub and target genes such as TP53, HRAS, MAPK11, RELA, IKZF3, IFNAR2, SKI, TNFRSF13C, JAK1, TRAF6, KLRF2, CD1A were identified from PPI network, target gene-miRNA regulatory network, and target gene-TF regulatory network. Study of the ROC showed that ten genes (CCL5, IFNAR2, JAK2, MX1, STAT1, BID, CD55, CD80, HAL-B, and HLA-DMA) were substantially involved in SARS-CoV-2 patients. The present investigation identified key genes and pathways that deepen our understanding of the molecular mechanisms of SARS-CoV-2 infection, and could be used for SARS-CoV-2 infection as diagnostic and therapeutic biomarkers.
Keywords: Bioinformatics analysis; Biomarkers; Differentially expressed genes; Protein–protein interaction (PPI) network; SARS-CoV-2 infection.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

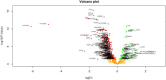
 ) represented up regulated significant genes and red dot on left side (
) represented up regulated significant genes and red dot on left side ( ) represented down regulated significant genes
) represented down regulated significant genes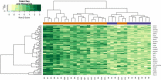

 ) denotes up regulated genes; Blue lines (
) denotes up regulated genes; Blue lines ( ) denotes edges (Interactions)
) denotes edges (Interactions)

 ) denotes down regulated genes; Pink lines (
) denotes down regulated genes; Pink lines ( ) denotes edges (Interactions)
) denotes edges (Interactions)
 ) denotes up regulated genes; Blue lines (
) denotes up regulated genes; Blue lines ( ) denotes edges (Interactions)
) denotes edges (Interactions)
 ) denotes down regulated genes; Pink lines (
) denotes down regulated genes; Pink lines ( ) denotes edges (Interactions)
) denotes edges (Interactions)
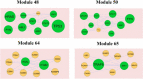
 ) are the up regulated genes; yellow diamond nodes (
) are the up regulated genes; yellow diamond nodes ( ) are the miRNAs; Pink lines (
) are the miRNAs; Pink lines ( ) denotes edges (Interactions)
) denotes edges (Interactions)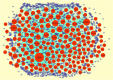
 ) are the down regulated genes; blue diamond nodes (
) are the down regulated genes; blue diamond nodes ( ) are the miRNAs; Sku blue lines (
) are the miRNAs; Sku blue lines ( ) denotes edges (Interactions)
) denotes edges (Interactions)
 ) are the up regulated genes; Blue triangle nodes (
) are the up regulated genes; Blue triangle nodes ( ) are the TFs; Purple line (
) are the TFs; Purple line ( ) denotes edges (Interactions)
) denotes edges (Interactions)
 ) are the down regulated genes; Blue triangle nodes (
) are the down regulated genes; Blue triangle nodes ( ) are the TFs; Yelow line (
) are the TFs; Yelow line ( ) denotes edges (Interactions)
) denotes edges (Interactions)
Similar articles
-
Bioinformatics analyses of significant genes, related pathways, and candidate diagnostic biomarkers and molecular targets in SARS-CoV-2/COVID-19.Gene Rep. 2020 Dec;21:100956. doi: 10.1016/j.genrep.2020.100956. Epub 2020 Nov 4. Gene Rep. 2020. PMID: 33553808 Free PMC article.
-
Potential Molecular Mechanisms and Remdesivir Treatment for Acute Respiratory Syndrome Corona Virus 2 Infection/COVID 19 Through RNA Sequencing and Bioinformatics Analysis.Bioinform Biol Insights. 2021 Dec 23;15:11779322211067365. doi: 10.1177/11779322211067365. eCollection 2021. Bioinform Biol Insights. 2021. PMID: 34992355 Free PMC article.
-
Identification of potential and novel target genes in pituitary prolactinoma by bioinformatics analysis.AIMS Neurosci. 2021 Feb 7;8(2):254-283. doi: 10.3934/Neuroscience.2021014. eCollection 2021. AIMS Neurosci. 2021. PMID: 33709028 Free PMC article.
-
Molecular mechanisms underlying gliomas and glioblastoma pathogenesis revealed by bioinformatics analysis of microarray data.Med Oncol. 2017 Sep 26;34(11):182. doi: 10.1007/s12032-017-1043-x. Med Oncol. 2017. PMID: 28952134
-
Exploration and validation of related hub gene expression during SARS-CoV-2 infection of human bronchial organoids.Hum Genomics. 2021 Mar 16;15(1):18. doi: 10.1186/s40246-021-00316-5. Hum Genomics. 2021. PMID: 33726831 Free PMC article.
Cited by
-
Predicted B Cell Epitopes Highlight the Potential for COVID-19 to Drive Self-Reactive Immunity.Front Bioinform. 2021 Aug 19;1:709533. doi: 10.3389/fbinf.2021.709533. eCollection 2021. Front Bioinform. 2021. PMID: 36303764 Free PMC article.
-
In silico Analyses of Immune System Protein Interactome Network, Single-Cell RNA Sequencing of Human Tissues, and Artificial Neural Networks Reveal Potential Therapeutic Targets for Drug Repurposing Against COVID-19.Front Pharmacol. 2021 Feb 26;12:598925. doi: 10.3389/fphar.2021.598925. eCollection 2021. Front Pharmacol. 2021. PMID: 33716737 Free PMC article.
-
Nucleic Acid-Based COVID-19 Therapy Targeting Cytokine Storms: Strategies to Quell the Storm.J Pers Med. 2022 Mar 3;12(3):386. doi: 10.3390/jpm12030386. J Pers Med. 2022. PMID: 35330388 Free PMC article. Review.
-
Epigenetic rewiring of pathways related to odour perception in immune cells exposed to SARS-CoV-2 in vivo and in vitro.Epigenetics. 2022 Dec;17(13):1875-1891. doi: 10.1080/15592294.2022.2089471. Epub 2022 Jun 26. Epigenetics. 2022. PMID: 35758003 Free PMC article.
-
H-Ras gene takes part to the host immune response to COVID-19.Cell Death Discov. 2021 Jun 25;7(1):158. doi: 10.1038/s41420-021-00541-w. Cell Death Discov. 2021. PMID: 34226505 Free PMC article. Review.
References
-
- Alaoui L, Palomino G, Zurawski S, Zurawski G, Coindre S, Dereuddre-Bosquet N, Lecuroux C, Goujard C, Vaslin B, Bourgeois C, et al. Early SIV and HIV infection promotes the LILRB2/MHC-I inhibitory axis in cDCs. Cell Mol Life Sci. 2018;75(10):1871–1887. doi: 10.1007/s00018-017-2712-9. - DOI - PMC - PubMed
-
- Antunes KH, Becker A, Franceschina C, de Freitas DD, Lape I, da Cunha MD, Leitão L, Rigo MM, Pinto LA, Stein RT, et al. Respiratory syncytial virus reduces STAT3 phosphorylation in human memory CD8 T cells stimulated with IL-21. Sci Rep. 2019;9(1):17766. doi: 10.1038/s41598-019-54240-9. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

