Key Molecules and Pathways Underlying Sporadic Amyotrophic Lateral Sclerosis: Integrated Analysis on Gene Expression Profiles of Motor Neurons
- PMID: 33240324
- PMCID: PMC7680998
- DOI: 10.3389/fgene.2020.578143
Key Molecules and Pathways Underlying Sporadic Amyotrophic Lateral Sclerosis: Integrated Analysis on Gene Expression Profiles of Motor Neurons
Abstract
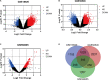
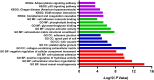
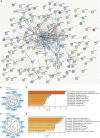
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder characterized by progressive loss of motor neurons. The complex mechanisms underlying ALS are yet to be elucidated, while the lack of disease biomarkers and therapeutic options are associated with the poor prognosis of ALS patients. In this study, we performed bioinformatics analysis to clarify potential mechanisms in sporadic ALS (sALS). We compared three gene expression profiles (GSE18920, GSE56500, and GSE68605) of motor neurons obtained from sALS patients and healthy controls to discover differentially expressed genes (DEGs), and then performed integrated bioinformatics analyses to identify key molecules and pathways underlying sALS. We found that these DEGs were mainly enriched in the structure and functions of extracellular matrix (ECM), while functional enrichment in blood vessel morphogenesis was less correlated with motor neurons. The clustered subnetworks of the constructed protein-protein interaction network for DEGs and the group of selected hub genes were more significantly involved in the organization of collagen-containing ECM. The transcriptional factors database proposed RelA and NF-κB1 from NF-κB family as the key regulators of these hub genes. These results mainly demonstrated the alternations in ECM-related gene expression in motor neurons and suggested the role of NF-κB regulatory pathway in the pathogenesis of sALS.
Keywords: FN1 gene; bioinformatics analysis; gene expression profiles; motor neurons; sporadic amyotrophic lateral sclerosis.
Copyright © 2020 Lin, Huang, Chen, Ye, Su and Yao.
Figures

Similar articles
-
Sporadic Amyotrophic Lateral Sclerosis Skeletal Muscle Transcriptome Analysis: A Comprehensive Examination of Differentially Expressed Genes.Biomolecules. 2024 Mar 20;14(3):377. doi: 10.3390/biom14030377. Biomolecules. 2024. PMID: 38540795 Free PMC article. Review.
-
Gene expression profiles and protein-protein interaction networks in amyotrophic lateral sclerosis patients with C9orf72 mutation.Orphanet J Rare Dis. 2016 Nov 5;11(1):148. doi: 10.1186/s13023-016-0531-y. Orphanet J Rare Dis. 2016. PMID: 27814735 Free PMC article.
-
A computational biology approach to identify potential protein biomarkers and drug targets for sporadic amyotrophic lateral sclerosis.Cell Signal. 2023 Dec;112:110915. doi: 10.1016/j.cellsig.2023.110915. Epub 2023 Oct 13. Cell Signal. 2023. PMID: 37838312
-
Network strategy to investigate differential pathways in sporadic amyotrophic lateral sclerosis.J Cancer Res Ther. 2018 Dec;14(Supplement):S1057-S1062. doi: 10.4103/0973-1482.199453. J Cancer Res Ther. 2018. PMID: 30539846
-
[Gene expression profile of spinal ventral horn in ALS].Brain Nerve. 2007 Oct;59(10):1129-39. Brain Nerve. 2007. PMID: 17969353 Review. Japanese.
Cited by
-
Quercetin targets VCAM1 to prevent diabetic cerebrovascular endothelial cell injury.Front Aging Neurosci. 2022 Sep 1;14:944195. doi: 10.3389/fnagi.2022.944195. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36118693 Free PMC article.
-
Proteomic analysis of cerebrospinal fluid of amyotrophic lateral sclerosis patients in the presence of autologous bone marrow derived mesenchymal stem cells.Stem Cell Res Ther. 2024 Sep 15;15(1):301. doi: 10.1186/s13287-024-03820-2. Stem Cell Res Ther. 2024. PMID: 39278909 Free PMC article. Clinical Trial.
-
Sporadic Amyotrophic Lateral Sclerosis Skeletal Muscle Transcriptome Analysis: A Comprehensive Examination of Differentially Expressed Genes.Biomolecules. 2024 Mar 20;14(3):377. doi: 10.3390/biom14030377. Biomolecules. 2024. PMID: 38540795 Free PMC article. Review.
-
Dysregulated Expression of Transposable Elements in TDP-43M337V Human Motor Neurons That Recapitulate Amyotrophic Lateral Sclerosis In Vitro.Int J Mol Sci. 2022 Dec 19;23(24):16222. doi: 10.3390/ijms232416222. Int J Mol Sci. 2022. PMID: 36555863 Free PMC article.
-
Key Disease Mechanisms Linked to Amyotrophic Lateral Sclerosis in Spinal Cord Motor Neurons.Front Mol Neurosci. 2022 Mar 15;15:825031. doi: 10.3389/fnmol.2022.825031. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35370543 Free PMC article.
References
-
- Brown R. H., Al-Chalabi A. (2017). Amyotrophic lateral sclerosis. N. Engl. J. Med. 377 162–172. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

