Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea
- PMID: 33234025
- PMCID: PMC7729029
- DOI: 10.1098/rsob.200302
Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea
Abstract
Phosphoglycerate kinase (PGK) is a glycolytic enzyme that is well conserved among the three domains of life. PGK is usually a monomeric enzyme of about 45 kDa that catalyses one of the two ATP-producing reactions in the glycolytic pathway, through the conversion of 1,3-bisphosphoglycerate (1,3BPGA) to 3-phosphoglycerate (3PGA). It also participates in gluconeogenesis, catalysing the opposite reaction to produce 1,3BPGA and ADP. Like most other glycolytic enzymes, PGK has also been catalogued as a moonlighting protein, due to its involvement in different functions not associated with energy metabolism, which include pathogenesis, interaction with nucleic acids, tumorigenesis progression, cell death and viral replication. In this review, we have highlighted the overall aspects of this enzyme, such as its structure, reaction kinetics, activity regulation and possible moonlighting functions in different protistan organisms, especially both free-living and parasitic Kinetoplastea. Our analysis of the genomes of different kinetoplastids revealed the presence of open-reading frames (ORFs) for multiple PGK isoforms in several species. Some of these ORFs code for unusually large PGKs. The products appear to contain additional structural domains fused to the PGK domain. A striking aspect is that some of these PGK isoforms are predicted to be catalytically inactive enzymes or 'dead' enzymes. The roles of PGKs in kinetoplastid parasites are analysed, and the apparent significance of the PGK gene duplication that gave rise to the different isoforms and their expression in Trypanosoma cruzi is discussed.
Keywords: Trypanosoma; domains; metabolism; moonlighting protein; phosphoglycerate kinase; protists.
Conflict of interest statement
We declare we have no competing interests.
Figures
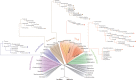
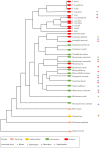
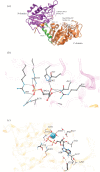


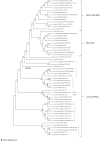
 , PGK domain having lost substrate (3PGA)-binding residues
, PGK domain having lost substrate (3PGA)-binding residues  , triosephosphate isomerase domain
, triosephosphate isomerase domain  , HTH domain
, HTH domain  , transmembrane segment
, transmembrane segment  , PAS domain
, PAS domain  , cyclic-nucleotide binding-domain
, cyclic-nucleotide binding-domain  . Import sequences and inserts identified in PGK isoenzymes through experimental and theoretical studies: putative PTS1
. Import sequences and inserts identified in PGK isoenzymes through experimental and theoretical studies: putative PTS1  , putative lysosome
, putative lysosome  , PGKA-like insert sequence
, PGKA-like insert sequence  .
.
Similar articles
-
Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.Structure. 1997 Nov 15;5(11):1475-83. doi: 10.1016/s0969-2126(97)00297-9. Structure. 1997. PMID: 9384563
-
Crystal structures of substrates and products bound to the phosphoglycerate kinase active site reveal the catalytic mechanism.Biochemistry. 1998 Mar 31;37(13):4429-36. doi: 10.1021/bi9724117. Biochemistry. 1998. PMID: 9521762
-
Molecular analysis of phosphoglycerate kinase in Trypanoplasma borreli and the evolution of this enzyme in kinetoplastida.Gene. 1998 Sep 14;217(1-2):91-9. doi: 10.1016/s0378-1119(98)00356-4. Gene. 1998. PMID: 9795157
-
Phosphoglycerate kinase.Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):93-104. doi: 10.1098/rstb.1981.0063. Philos Trans R Soc Lond B Biol Sci. 1981. PMID: 6115427 Review.
-
Flexibility and folding of phosphoglycerate kinase.Biochimie. 1990 Jun-Jul;72(6-7):417-29. doi: 10.1016/0300-9084(90)90066-p. Biochimie. 1990. PMID: 2124145 Review.
Cited by
-
Integrity assay for messenger RNA in mouse and human brain samples and synaptosomal preparations.iScience. 2024 Jun 29;27(8):110419. doi: 10.1016/j.isci.2024.110419. eCollection 2024 Aug 16. iScience. 2024. PMID: 39108710 Free PMC article.
-
Regulation of Autophagy via Carbohydrate and Lipid Metabolism in Cancer.Cancers (Basel). 2023 Apr 7;15(8):2195. doi: 10.3390/cancers15082195. Cancers (Basel). 2023. PMID: 37190124 Free PMC article. Review.
-
Transcriptomic analysis of the response of Avena sativa to Bacillus amyloliquefaciens DGL1.Front Microbiol. 2024 Apr 3;15:1321989. doi: 10.3389/fmicb.2024.1321989. eCollection 2024. Front Microbiol. 2024. PMID: 38633698 Free PMC article.
-
BONCAT-iTRAQ Labelling Reveals Molecular Markers of Adaptive Responses in Toxoplasma gondii to Pyrimethamine Treatment.Pathogens. 2024 Oct 8;13(10):879. doi: 10.3390/pathogens13100879. Pathogens. 2024. PMID: 39452750 Free PMC article.
-
Analysis of Hyperosmotic Tolerance Mechanisms in Gracilariopsis lemaneiformis Based on Weighted Co-Expression Network Analysis.Genes (Basel). 2024 Jun 13;15(6):781. doi: 10.3390/genes15060781. Genes (Basel). 2024. PMID: 38927717 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

