An immune-based biomarker signature is associated with mortality in COVID-19 patients
- PMID: 33232303
- PMCID: PMC7821609
- DOI: 10.1172/jci.insight.144455
An immune-based biomarker signature is associated with mortality in COVID-19 patients
Abstract
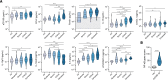
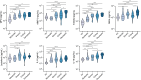
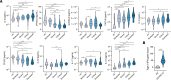
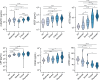
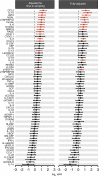
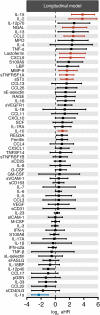
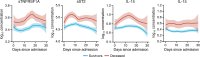
Immune and inflammatory responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) contribute to disease severity of coronavirus disease 2019 (COVID-19). However, the utility of specific immune-based biomarkers to predict clinical outcome remains elusive. Here, we analyzed levels of 66 soluble biomarkers in 175 Italian patients with COVID-19 ranging from mild/moderate to critical severity and assessed type I IFN-, type II IFN-, and NF-κB-dependent whole-blood transcriptional signatures. A broad inflammatory signature was observed, implicating activation of various immune and nonhematopoietic cell subsets. Discordance between IFN-α2a protein and IFNA2 transcript levels in blood suggests that type I IFNs during COVID-19 may be primarily produced by tissue-resident cells. Multivariable analysis of patients' first samples revealed 12 biomarkers (CCL2, IL-15, soluble ST2 [sST2], NGAL, sTNFRSF1A, ferritin, IL-6, S100A9, MMP-9, IL-2, sVEGFR1, IL-10) that when increased were independently associated with mortality. Multivariate analyses of longitudinal biomarker trajectories identified 8 of the aforementioned biomarkers (IL-15, IL-2, NGAL, CCL2, MMP-9, sTNFRSF1A, sST2, IL-10) and 2 additional biomarkers (lactoferrin, CXCL9) that were substantially associated with mortality when increased, while IL-1α was associated with mortality when decreased. Among these, sST2, sTNFRSF1A, IL-10, and IL-15 were consistently higher throughout the hospitalization in patients who died versus those who recovered, suggesting that these biomarkers may provide an early warning of eventual disease outcome.
Keywords: COVID-19; Chemokines; Cytokines; Immunology.
Conflict of interest statement
Figures

Similar articles
-
Interplay of Antibody and Cytokine Production Reveals CXCL13 as a Potential Novel Biomarker of Lethal SARS-CoV-2 Infection.mSphere. 2021 Jan 20;6(1):e01324-20. doi: 10.1128/mSphere.01324-20. mSphere. 2021. PMID: 33472985 Free PMC article.
-
Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease.JCI Insight. 2020 Jul 9;5(13):e139834. doi: 10.1172/jci.insight.139834. JCI Insight. 2020. PMID: 32501293 Free PMC article.
-
Cytokine Profiles Associated With Worse Prognosis in a Hospitalized Peruvian COVID-19 Cohort.Front Immunol. 2021 Sep 1;12:700921. doi: 10.3389/fimmu.2021.700921. eCollection 2021. Front Immunol. 2021. PMID: 34539631 Free PMC article.
-
Disruption of CCR5 signaling to treat COVID-19-associated cytokine storm: Case series of four critically ill patients treated with leronlimab.J Transl Autoimmun. 2021;4:100083. doi: 10.1016/j.jtauto.2021.100083. Epub 2021 Jan 6. J Transl Autoimmun. 2021. PMID: 33521616 Free PMC article. Review.
-
Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis.Clin Microbiol Rev. 2021 May 12;34(3):e00299-20. doi: 10.1128/CMR.00299-20. Print 2021 Jun 16. Clin Microbiol Rev. 2021. PMID: 33980688 Free PMC article. Review.
Cited by
-
The Intersection of COVID-19 and Autoimmunity: What is Our Current Understanding?Pathog Immun. 2021 Mar 8;6(1):31-54. doi: 10.20411/pai.v6i1.417. eCollection 2021. Pathog Immun. 2021. PMID: 33969248 Free PMC article.
-
Role of cystatin C and calprotectin as potential early prognostic biomarkers in COVID-19 patients admitted to a dedicated COVID care facility.J Family Med Prim Care. 2022 Jul;11(7):3971-3979. doi: 10.4103/jfmpc.jfmpc_545_22. Epub 2022 Jul 22. J Family Med Prim Care. 2022. PMID: 36387740 Free PMC article.
-
The Association of Low CD4 Expression on Monocytes and Low CD8+ T-Cell Count at Hospital Admission Predicts the Need for Mechanical Ventilation in Patients With COVID-19 Pneumonia: A Prospective Monocentric Cohort Study.Crit Care Explor. 2022 Dec 7;4(12):e0810. doi: 10.1097/CCE.0000000000000810. eCollection 2022 Dec. Crit Care Explor. 2022. PMID: 36518218 Free PMC article.
-
The immune response as a double-edged sword: The lesson learnt during the COVID-19 pandemic.Immunology. 2022 Nov;167(3):287-302. doi: 10.1111/imm.13564. Epub 2022 Sep 5. Immunology. 2022. PMID: 35971810 Free PMC article. Review.
-
Immune Senescence, Immunosenescence and Aging.Front Aging. 2022 May 30;3:900028. doi: 10.3389/fragi.2022.900028. eCollection 2022. Front Aging. 2022. PMID: 35821850 Free PMC article. Review.
References
-
- John Hopkins Coronavirus Resource Center. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html Updated November 30, 2020. Accessed November 30, 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

