The expanding world of plant J-domain proteins
- PMID: 33223602
- PMCID: PMC7678915
- DOI: 10.1080/07352689.2019.1693716
The expanding world of plant J-domain proteins
Abstract
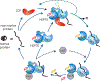
Plants maintain cellular proteostasis during different phases of growth and development despite a barrage of biotic and abiotic stressors in an ever-changing environment. This requires a collaborative effort of a cadre of molecular chaperones. Hsp70s and their obligate co-chaperones, J-domain proteins (JDPs), are arguably the most ubiquitous and formidable components of the cellular chaperone network, facilitating numerous and diverse cellular processes and allowing survival under a plethora of stressful conditions. JDPs are also among the most versatile chaperones. Compared to Hsp70s, the number of JDP-encoding genes has proliferated, suggesting the emergence of highly complex Hsp70-JDP networks, particularly in plants. Recent studies indicate that besides the increase in the number of JDP encoding genes; regulatory differences, neo- and sub-functionalization, and inter- and intra-class combinatorial interactions, is rapidly expanding the repertoire of Hsp70-JDP systems. This results in highly robust and functionally diverse chaperone networks in plants. Here, we review the current status of plant JDP research and discuss how the paradigm shift in the field can be exploited toward a better understanding of JDP function and evolution.
Keywords: Evolution; Hsp40; Hsp70; JDP; plants.
Figures




Similar articles
-
J-like protein family of Arabidopsis thaliana: the enigmatic cousins of J-domain proteins.Plant Cell Rep. 2022 Jun;41(6):1343-1355. doi: 10.1007/s00299-022-02857-y. Epub 2022 Mar 15. Plant Cell Rep. 2022. PMID: 35290497 Review.
-
J-domain protein chaperone circuits in proteostasis and disease.Trends Cell Biol. 2023 Jan;33(1):30-47. doi: 10.1016/j.tcb.2022.05.004. Epub 2022 Jun 18. Trends Cell Biol. 2023. PMID: 35729039 Free PMC article. Review.
-
Data-driven large-scale genomic analysis reveals an intricate phylogenetic and functional landscape in J-domain proteins.Proc Natl Acad Sci U S A. 2023 Aug 8;120(32):e2218217120. doi: 10.1073/pnas.2218217120. Epub 2023 Jul 31. Proc Natl Acad Sci U S A. 2023. PMID: 37523524 Free PMC article. Review.
-
The Hsp70 and JDP proteins: Structure-function perspective on molecular chaperone activity.Enzymes. 2023;54:221-245. doi: 10.1016/bs.enz.2023.07.008. Epub 2023 Sep 29. Enzymes. 2023. PMID: 37945173 Review.
-
Extracellular chaperone networks and the export of J-domain proteins.J Biol Chem. 2023 Feb;299(2):102840. doi: 10.1016/j.jbc.2022.102840. Epub 2022 Dec 26. J Biol Chem. 2023. PMID: 36581212 Free PMC article. Review.
Cited by
-
J-like protein family of Arabidopsis thaliana: the enigmatic cousins of J-domain proteins.Plant Cell Rep. 2022 Jun;41(6):1343-1355. doi: 10.1007/s00299-022-02857-y. Epub 2022 Mar 15. Plant Cell Rep. 2022. PMID: 35290497 Review.
-
Arabidopsis Dph4 is an Hsp70 Cochaperone with Iron-Binding Properties.ACS Omega. 2024 Aug 28;9(36):37650-37661. doi: 10.1021/acsomega.4c01776. eCollection 2024 Sep 10. ACS Omega. 2024. PMID: 39281955 Free PMC article.
-
J-domain protein chaperone circuits in proteostasis and disease.Trends Cell Biol. 2023 Jan;33(1):30-47. doi: 10.1016/j.tcb.2022.05.004. Epub 2022 Jun 18. Trends Cell Biol. 2023. PMID: 35729039 Free PMC article. Review.
-
Genome-wide profiling of histone (H3) lysine 4 (K4) tri-methylation (me3) under drought, heat, and combined stresses in switchgrass.BMC Genomics. 2024 Feb 29;25(1):223. doi: 10.1186/s12864-024-10068-w. BMC Genomics. 2024. PMID: 38424499 Free PMC article.
-
Drought Stress Amelioration Attributes of Plant-Associated Microbiome on Agricultural Plants.Bioinform Biol Insights. 2024 Mar 8;18:11779322241233442. doi: 10.1177/11779322241233442. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 38464334 Free PMC article. Review.
References
-
- Bekh-Ochir D, Shimada S, Yamagami A, Kanda S, Ogawa K, Nakazawa M, Matsui M, Sakuta M, Osada H, Asami T, and Nakano T 2013. A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 237: 1509–1525. doi:10.1007/s00425-013-1859-3 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
