lnc-MAP3K13-7:1 Inhibits Ovarian GC Proliferation in PCOS via DNMT1 Downregulation-Mediated CDKN1A Promoter Hypomethylation
- PMID: 33212300
- PMCID: PMC7934583
- DOI: 10.1016/j.ymthe.2020.11.018
lnc-MAP3K13-7:1 Inhibits Ovarian GC Proliferation in PCOS via DNMT1 Downregulation-Mediated CDKN1A Promoter Hypomethylation
Abstract
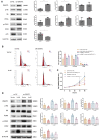
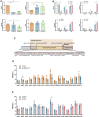
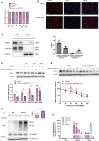
Polycystic ovary syndrome (PCOS) is an endocrine-related disease and global cause of infertility that is associated with abnormal folliculogenesis. Inhibited granulosa cell (GC) proliferation is recognized as a key factor that underlies aberrant follicle maturation. Many epigenetic landscape modifications have been characterized in PCOS patients. However, the epigenetic regulation pathways in follicular dysplasia are not completely understood. In this study, we reported a novel mechanism of DNA hypomethylation induced by long non-coding RNAs (lncRNAs) and its function in cell cycle progression. We observed that lnc-MAP3K13-7:1 was highly expressed in GCs from patients with PCOS, with concomitant global DNA hypomethylation, decreased DNA methyltransferase 1 (DNMT1) expression, and increased cyclin-dependent kinase inhibitor 1A (CDKN1A, p21) expression. In KGN cells, lnc-MAP3K13-7:1 overexpression resulted in cell cycle arrest in the G0/G1 phase, as well as the molecular inhibition and genetic silencing of DNMT1. Mechanistically, lnc-MAP3K13-7:1 inhibited DNMT1 expression by acting as a protein-binding scaffold and inducing ubiquitin-mediated DNMT1 protein degradation. Moreover, DNMT1-dependent CDKN1A promoter hypomethylation increased CDKN1A transcription, resulting in attenuated GC growth. Our work uncovered a novel and essential mechanism through which lnc-MAP3K13-7:1-dependent DNMT1 inhibition regulates CDKN1A/p21 expression and inhibits GC proliferation.
Keywords: DNA methylation; DNMT1; PCOS; epigenetics; granulosa cell proliferation; lncRNA.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome.J Clin Endocrinol Metab. 2015 May;100(5):E729-38. doi: 10.1210/jc.2014-3827. Epub 2015 Feb 19. J Clin Endocrinol Metab. 2015. PMID: 25695884 Free PMC article.
-
Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway.Mol Hum Reprod. 2019 Jan 1;25(1):17-29. doi: 10.1093/molehr/gay045. Mol Hum Reprod. 2019. PMID: 30371869
-
Downregulation of Lnc-OC1 attenuates the pathogenesis of polycystic ovary syndrome.Mol Cell Endocrinol. 2020 Apr 15;506:110760. doi: 10.1016/j.mce.2020.110760. Epub 2020 Feb 15. Mol Cell Endocrinol. 2020. PMID: 32070768
-
Long non-coding RNAs in ovarian granulosa cells.J Ovarian Res. 2020 Jun 5;13(1):63. doi: 10.1186/s13048-020-00663-2. J Ovarian Res. 2020. PMID: 32503679 Free PMC article. Review.
-
Long noncoding RNAs as a piece of polycystic ovary syndrome puzzle.Mol Biol Rep. 2021 Apr;48(4):3845-3851. doi: 10.1007/s11033-021-06196-1. Epub 2021 May 15. Mol Biol Rep. 2021. PMID: 33993404 Review.
Cited by
-
Roles of Noncoding RNA in Reproduction.Front Genet. 2021 Dec 9;12:777510. doi: 10.3389/fgene.2021.777510. eCollection 2021. Front Genet. 2021. PMID: 34956326 Free PMC article. Review.
-
Integrated bioinformatics analysis elucidates granulosa cell whole-transcriptome landscape of PCOS in China.J Ovarian Res. 2023 Aug 3;16(1):154. doi: 10.1186/s13048-023-01223-0. J Ovarian Res. 2023. PMID: 37537636 Free PMC article.
-
LncRNA SNHG12 promotes cell proliferation and inhibits apoptosis of granulosa cells in polycystic ovarian syndrome by sponging miR-129 and miR-125b.J Ovarian Res. 2024 Apr 2;17(1):72. doi: 10.1186/s13048-024-01392-6. J Ovarian Res. 2024. PMID: 38566229 Free PMC article.
-
ER stress-induced LINC00173 promotes the apoptosis of ovarian granulosa cells by regulating the HRK/PI3K/AKT pathway in polycystic ovary syndrome.Sci Rep. 2024 Oct 20;14(1):24636. doi: 10.1038/s41598-024-75178-7. Sci Rep. 2024. PMID: 39428498 Free PMC article.
-
LncRNA-AK137033 inhibits the osteogenic potential of adipose-derived stem cells in diabetic osteoporosis by regulating Wnt signaling pathway via DNA methylation.Cell Prolif. 2022 Jan;55(1):e13174. doi: 10.1111/cpr.13174. Epub 2021 Dec 24. Cell Prolif. 2022. PMID: 34953002 Free PMC article.
References
-
- Bozdag G., Mumusoglu S., Zengin D., Karabulut E., Yildiz B.O. The prevalence and phenotypic features of polycystic ovary syndrome: a systematic review and meta-analysis. Hum. Reprod. 2016;31:2841–2855. - PubMed
-
- Skiba M.A., Islam R.M., Bell R.J., Davis S.R. Understanding variation in prevalence estimates of polycystic ovary syndrome: a systematic review and meta-analysis. Hum. Reprod. Update. 2018;24:694–709. - PubMed
-
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil. Steril. 2004;81:19–25. - PubMed
-
- Franks S., Stark J., Hardy K. Follicle dynamics and anovulation in polycystic ovary syndrome. Hum. Reprod. Update. 2008;14:367–378. - PubMed
-
- Webber L.J., Stubbs S., Stark J., Trew G.H., Margara R., Hardy K., Franks S. Formation and early development of follicles in the polycystic ovary. Lancet. 2003;362:1017–1021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

