The Early Evolution of Oral Poliovirus Vaccine Is Shaped by Strong Positive Selection and Tight Transmission Bottlenecks
- PMID: 33212020
- PMCID: PMC7815045
- DOI: 10.1016/j.chom.2020.10.011
The Early Evolution of Oral Poliovirus Vaccine Is Shaped by Strong Positive Selection and Tight Transmission Bottlenecks
Abstract
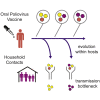
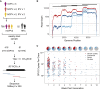
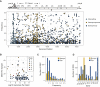
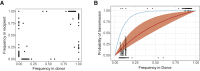
The emergence of circulating vaccine-derived polioviruses through evolution of the oral polio vaccine (OPV) poses a significant obstacle to polio eradication. Understanding the early genetic changes that occur as OPV evolves and transmits is important for preventing future outbreaks. Here, we use deep sequencing to define the evolutionary trajectories of type 2 OPV in a vaccine trial. By sequencing 497 longitudinal stool samples from 271 OPV2 recipients and household contacts, we were able to examine the extent of convergent evolution in vaccinated individuals and the amount of viral diversity that is transmitted. In addition to rapid reversion of key attenuating mutations, we identify strong selection at 19 sites across the genome. We find that a tight transmission bottleneck limits the onward transmission of these early adaptive mutations. Our results highlight the distinct evolutionary dynamics of live attenuated virus vaccines and have important implications for the success of next-generation OPV.
Keywords: evolution; live vaccines; poliovirus; sequencing; transmission bottlenecks; vaccine evolution.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


Comment in
-
Mutation, Selection, and Bottlenecks in Polio Vaccine Reversion.Cell Host Microbe. 2021 Jan 13;29(1):3-5. doi: 10.1016/j.chom.2020.12.018. Cell Host Microbe. 2021. PMID: 33444554
Similar articles
-
The safety and immunogenicity of two novel live attenuated monovalent (serotype 2) oral poliovirus vaccines in healthy adults: a double-blind, single-centre phase 1 study.Lancet. 2019 Jul 13;394(10193):148-158. doi: 10.1016/S0140-6736(19)31279-6. Epub 2019 Jun 4. Lancet. 2019. PMID: 31174831 Free PMC article. Clinical Trial.
-
Genetic stabilization of attenuated oral vaccines against poliovirus types 1 and 3.Nature. 2023 Jul;619(7968):135-142. doi: 10.1038/s41586-023-06212-3. Epub 2023 Jun 14. Nature. 2023. PMID: 37316671 Free PMC article.
-
Molecular evolution of oral poliovirus vaccine strains during multiplication in humans and possible implications for global eradication of poliovirus.Acta Virol. 2000 Apr;44(2):109-17. Acta Virol. 2000. PMID: 10989702 Review.
-
A Cluster of Paralytic Poliomyelitis Cases Due to Transmission of Slightly Diverged Sabin 2 Vaccine Poliovirus.J Virol. 2016 Jun 10;90(13):5978-88. doi: 10.1128/JVI.00277-16. Print 2016 Jul 1. J Virol. 2016. PMID: 27099315 Free PMC article.
-
Risks associated with the use of live-attenuated vaccine poliovirus strains and the strategies for control and eradication of paralytic poliomyelitis.Expert Rev Vaccines. 2012 May;11(5):609-28. doi: 10.1586/erv.12.28. Expert Rev Vaccines. 2012. PMID: 22827246 Review.
Cited by
-
Review of Poliovirus Transmission and Economic Modeling to Support Global Polio Eradication: 2020-2024.Pathogens. 2024 May 22;13(6):435. doi: 10.3390/pathogens13060435. Pathogens. 2024. PMID: 38921733 Free PMC article. Review.
-
Picornavirus 3C Proteins Intervene in Host Cell Processes through Proteolysis and Interactions with RNA.Viruses. 2023 Dec 12;15(12):2413. doi: 10.3390/v15122413. Viruses. 2023. PMID: 38140654 Free PMC article. Review.
-
Rapid emergence and transmission of virulence-associated mutations in the oral poliovirus vaccine following vaccination campaigns.NPJ Vaccines. 2023 Sep 25;8(1):137. doi: 10.1038/s41541-023-00740-9. NPJ Vaccines. 2023. PMID: 37749086 Free PMC article.
-
The development of resistance to an inhibitor of a cellular protein reveals a critical interaction between the enterovirus protein 2C and a small GTPase Arf1.PLoS Pathog. 2023 Sep 18;19(9):e1011673. doi: 10.1371/journal.ppat.1011673. eCollection 2023 Sep. PLoS Pathog. 2023. PMID: 37721955 Free PMC article.
-
SARS-CoV-2 shedding and evolution in immunocompromised hosts during the Omicron period: a multicenter prospective analysis.medRxiv [Preprint]. 2023 Aug 24:2023.08.22.23294416. doi: 10.1101/2023.08.22.23294416. medRxiv. 2023. Update in: Lancet Microbe. 2024 Mar;5(3):e235-e246. doi: 10.1016/S2666-5247(23)00336-1. PMID: 37662226 Free PMC article. Updated. Preprint.
References
-
- Alam N., Ali T., Razzaque A., Rahman M., Zahirul Haq M., Saha S.K., Ahmed A., Sarder A.M., Moinuddin Haider M., Yunus M. Health and demographic surveillance system (HDSS) in Matlab, Bangladesh. Int. J. Epidemiol. 2017;46:809–816. - PubMed
-
- Boot H.J., Sonsma J., van Nunen F., Abbink F., Kimman T.G., Buisman A.M. Determinants of monovalent oral polio vaccine mutagenesis in vaccinated elderly people. Vaccine. 2007;25:4706–4714. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

