Dual Effects of Let-7b in the Early Stage of Hepatitis C Virus Infection
- PMID: 33208444
- PMCID: PMC7851561
- DOI: 10.1128/JVI.01800-20
Dual Effects of Let-7b in the Early Stage of Hepatitis C Virus Infection
Abstract
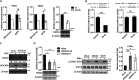
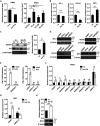
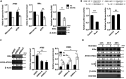
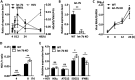
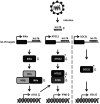
MicroRNA let-7b expression is induced by infection of hepatitis C virus (HCV) and is involved in the regulation of HCV replication by directly targeting the HCV genome. The current study demonstrated that let-7b directly targets negative regulators of type I interferon (IFN) signaling thereby limiting HCV replication in the early stage of HCV infection. Let-7b-regulated genes which are involved in host cellular responses to HCV infection were unveiled by microarray profiling and bioinformatic analyses, followed by various molecular and cellular assays using Huh7 cells expressing wild-type (WT) or the seed region-mutated let-7b. Let-7b targeted the cytokine signaling 1 (SOCS1) protein, a negative regulator of JAK/STAT signaling, which then enhanced STAT1-Y701 phosphorylation leading to increased expression of the downstream interferon-stimulated genes (ISGs). Let-7b augmented retinoic acid-inducible gene I (RIG-I) signaling, but not MDA5, to phosphorylate and nuclear translocate IRF3 leading to increased expression of IFN-β. Let-7b directly targeted the ATG12 and IκB kinase alpha (IKKα) transcripts and reduced the interaction of the ATG5-ATG12 conjugate and RIG-I leading to increased expression of IFN, which may further stimulate JAK/STAT signaling. Let-7b induced by HCV infection elicits dual effects on IFN expression and signaling, along with targeting the coding sequences of NS5B and 5' UTR of the HCV genome, and limits HCV RNA accumulation in the early stage of HCV infection. Controlling let-7b expression is thereby crucial in the intervention of HCV infection.IMPORTANCE HCV is a leading cause of liver disease, with an estimated 71 million people infected worldwide. During HCV infection, type I interferon (IFN) signaling displays potent antiviral and immunomodulatory effects. Host factors, including microRNAs (miRNAs), play a role in upregulating IFN signaling to limit HCV replication. Let-7b is a liver-abundant miRNA that is induced by HCV infection and targets the HCV genome to suppress HCV RNA accumulation. In this study, we demonstrated that let-7b, as a positive regulator of type I IFN signaling, plays dual roles against HCV replication by increasing the expression of IFN and interferon-sensitive response element (ISRE)-driven interferon-stimulated genes (ISGs) in the early stage of HCV infection. This study sheds new insight into understanding the role of let-7b in combatting HCV infection. Clarifying IFN signaling regulated by miRNA during the early phase of HCV infection may help researchers understand the initial defense mechanisms to other RNA viruses.
Keywords: hepatitis C virus; interferon; let-7b.
Copyright © 2021 American Society for Microbiology.
Figures

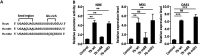


Similar articles
-
Let-7b is a novel regulator of hepatitis C virus replication.Cell Mol Life Sci. 2012 Aug;69(15):2621-33. doi: 10.1007/s00018-012-0940-6. Epub 2012 Mar 6. Cell Mol Life Sci. 2012. PMID: 22391672 Free PMC article.
-
High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1.J Virol. 2013 Sep;87(17):9707-18. doi: 10.1128/JVI.00802-13. Epub 2013 Jul 3. J Virol. 2013. PMID: 23824794 Free PMC article.
-
Effect of ethanol on innate antiviral pathways and HCV replication in human liver cells.Virol J. 2005 Dec 2;2:89. doi: 10.1186/1743-422X-2-89. Virol J. 2005. PMID: 16324217 Free PMC article.
-
Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review.
-
Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection.Cell Mol Immunol. 2016 Jan;13(1):11-35. doi: 10.1038/cmi.2014.127. Epub 2014 Dec 29. Cell Mol Immunol. 2016. PMID: 25544499 Free PMC article. Review.
Cited by
-
Dual Effects of 3-epi-betulin from Daphniphyllum glaucescens in Suppressing SARS-CoV-2-Induced Inflammation and Inhibiting Virus Entry.Int J Mol Sci. 2023 Dec 1;24(23):17040. doi: 10.3390/ijms242317040. Int J Mol Sci. 2023. PMID: 38069363 Free PMC article.
-
Host combats porcine reproductive and respiratory syndrome virus infection at non-coding RNAs level.Virulence. 2024 Dec;15(1):2416551. doi: 10.1080/21505594.2024.2416551. Epub 2024 Oct 18. Virulence. 2024. PMID: 39403796 Free PMC article. Review.
-
Role of microRNAs in host defense against porcine reproductive and respiratory syndrome virus infection: a hidden front line.Front Immunol. 2024 Mar 25;15:1376958. doi: 10.3389/fimmu.2024.1376958. eCollection 2024. Front Immunol. 2024. PMID: 38590524 Free PMC article. Review.
-
Micro-Players of Great Significance-Host microRNA Signature in Viral Infections in Humans and Animals.Int J Mol Sci. 2022 Sep 11;23(18):10536. doi: 10.3390/ijms231810536. Int J Mol Sci. 2022. PMID: 36142450 Free PMC article. Review.
-
MicroRNA let-7 and viral infections: focus on mechanisms of action.Cell Mol Biol Lett. 2022 Feb 14;27(1):14. doi: 10.1186/s11658-022-00317-9. Cell Mol Biol Lett. 2022. PMID: 35164678 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

