The RNA Polymerase α Subunit Recognizes the DNA Shape of the Upstream Promoter Element
- PMID: 33205945
- PMCID: PMC8100869
- DOI: 10.1021/acs.biochem.0c00571
The RNA Polymerase α Subunit Recognizes the DNA Shape of the Upstream Promoter Element
Abstract
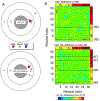
We demonstrate here that the α subunit C-terminal domain of Escherichia coli RNA polymerase (αCTD) recognizes the upstream promoter (UP) DNA element via its characteristic minor groove shape and electrostatic potential. In two compositionally distinct crystallized assemblies, a pair of αCTD subunits bind in tandem to the UP element consensus A-tract that is 6 bp in length (A6-tract), each with their arginine 265 guanidinium group inserted into the minor groove. The A6-tract minor groove is significantly narrowed in these crystal structures, as well as in computationally predicted structures of free and bound DNA duplexes derived by Monte Carlo and molecular dynamics simulations, respectively. The negative electrostatic potential of free A6-tract DNA is substantially enhanced compared to that of generic DNA. Shortening the A-tract by 1 bp is shown to "knock out" binding of the second αCTD through widening of the minor groove. Furthermore, in computationally derived structures with arginine 265 mutated to alanine in either αCTD, either with or without the "knockout" DNA mutation, contact with the DNA is perturbed, highlighting the importance of arginine 265 in achieving αCTD-DNA binding. These results demonstrate that the importance of the DNA shape in sequence-dependent recognition of DNA by RNA polymerase is comparable to that of certain transcription factors.
Conflict of interest statement
Figures

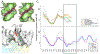
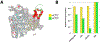
Similar articles
-
The C-terminal domains of the RNA polymerase alpha subunits: contact site with Fis and localization during co-activation with CRP at the Escherichia coli proP P2 promoter.J Mol Biol. 2002 Feb 22;316(3):517-29. doi: 10.1006/jmbi.2001.5391. J Mol Biol. 2002. PMID: 11866515
-
Novel protein--protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase alpha subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress.J Mol Biol. 2004 Oct 22;343(3):513-32. doi: 10.1016/j.jmb.2004.08.057. J Mol Biol. 2004. PMID: 15465042
-
Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase alpha subunits.J Mol Biol. 1998 Apr 10;277(4):789-804. doi: 10.1006/jmbi.1998.1666. J Mol Biol. 1998. PMID: 9545373
-
Transcription activation at Class I CAP-dependent promoters.Mol Microbiol. 1993 May;8(5):797-802. doi: 10.1111/j.1365-2958.1993.tb01626.x. Mol Microbiol. 1993. PMID: 8394979 Review.
-
Activation of P2 late transcription by P2 Ogr protein requires a discrete contact site on the C terminus of the alpha subunit of Escherichia coli RNA polymerase.J Mol Biol. 1997 Nov 21;274(1):1-7. doi: 10.1006/jmbi.1997.1390. J Mol Biol. 1997. PMID: 9398509 Review.
Cited by
-
Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation.Phage (New Rochelle). 2021 Dec 1;2(4):170-182. doi: 10.1089/phage.2021.0013. Epub 2021 Dec 16. Phage (New Rochelle). 2021. PMID: 35083439 Free PMC article.
-
Elucidating the biology of transcription factor-DNA interaction for accurate identification of cis-regulatory elements.Curr Opin Plant Biol. 2022 Aug;68:102232. doi: 10.1016/j.pbi.2022.102232. Epub 2022 Jun 6. Curr Opin Plant Biol. 2022. PMID: 35679803 Free PMC article. Review.
-
Delineation of the DNA Structural Features of Eukaryotic Core Promoter Classes.ACS Omega. 2022 Feb 9;7(7):5657-5669. doi: 10.1021/acsomega.1c04603. eCollection 2022 Feb 22. ACS Omega. 2022. PMID: 35224327 Free PMC article.
-
Structural basis of ribosomal RNA transcription regulation.Nat Commun. 2021 Jan 22;12(1):528. doi: 10.1038/s41467-020-20776-y. Nat Commun. 2021. PMID: 33483500 Free PMC article.
-
Insight into the sequence-specific elements leading to increased DNA bending and ligase-mediated circularization propensity by antitumor trabectedin.J Comput Aided Mol Des. 2021 Jun;35(6):707-719. doi: 10.1007/s10822-021-00396-4. Epub 2021 Jun 9. J Comput Aided Mol Des. 2021. PMID: 34105031
References
-
- Ebright RH; Busby S (1995) The Escherichia coli RNA polymerase alpha subunit: structure and function, Current opinion in genetics & development 5, 197–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

