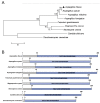
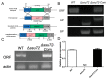
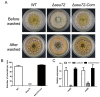
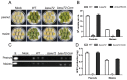
Ssu72 Regulates Fungal Development, Aflatoxin Biosynthesis and Pathogenicity in Aspergillus flavus
- PMID: 33202955
- PMCID: PMC7696088
- DOI: 10.3390/toxins12110717
Ssu72 Regulates Fungal Development, Aflatoxin Biosynthesis and Pathogenicity in Aspergillus flavus
Abstract
The RNA polymerase II (Pol II) transcription process is coordinated by the reversible phosphorylation of its largest subunit-carboxy terminal domain (CTD). Ssu72 is identified as a CTD phosphatase with specificity for phosphorylation of Ser5 and Ser7 and plays critical roles in regulation of transcription cycle in eukaryotes. However, the biofunction of Ssu72 is still unknown in Aspergillus flavus, which is a plant pathogenic fungus and produces one of the most toxic mycotoxins-aflatoxin. Here, we identified a putative phosphatase Ssu72 and investigated the function of Ssu72 in A. flavus. Deletion of ssu72 resulted in severe defects in vegetative growth, conidiation and sclerotia formation. Additionally, we found that phosphatase Ssu72 positively regulates aflatoxin production through regulating expression of aflatoxin biosynthesis cluster genes. Notably, seeds infection assays indicated that phosphatase Ssu72 is crucial for pathogenicity of A. flavus. Furthermore, the Δssu72 mutant exhibited more sensitivity to osmotic and oxidative stresses. Taken together, our study suggests that the putative phosphatase Ssu72 is involved in fungal development, aflatoxin production and pathogenicity in A. flavus, and may provide a novel strategy to prevent the contamination of this pathogenic fungus.
Keywords: A. flavus; aflatoxins; pathogenicity; phosphatase; ssu72.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The Putative Histone Methyltransferase DOT1 Regulates Aflatoxin and Pathogenicity Attributes in Aspergillus flavus.Toxins (Basel). 2017 Jul 24;9(7):232. doi: 10.3390/toxins9070232. Toxins (Basel). 2017. PMID: 28737735 Free PMC article.
-
The Aspergillus flavus Phosphatase CDC14 Regulates Development, Aflatoxin Biosynthesis and Pathogenicity.Front Cell Infect Microbiol. 2018 May 7;8:141. doi: 10.3389/fcimb.2018.00141. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29868497 Free PMC article.
-
The target of rapamycin signaling pathway regulates vegetative development, aflatoxin biosynthesis, and pathogenicity in Aspergillus flavus.Elife. 2024 Jul 11;12:RP89478. doi: 10.7554/eLife.89478. Elife. 2024. PMID: 38990939 Free PMC article.
-
Understanding the genetics of regulation of aflatoxin production and Aspergillus flavus development.Mycopathologia. 2006 Sep;162(3):155-66. doi: 10.1007/s11046-006-0050-9. Mycopathologia. 2006. PMID: 16944283 Review.
-
Aspergillus flavus.Annu Rev Phytopathol. 2011;49:107-33. doi: 10.1146/annurev-phyto-072910-095221. Annu Rev Phytopathol. 2011. PMID: 21513456 Review.
Cited by
-
Ssu72: a versatile protein with functions in transcription and beyond.Front Mol Biosci. 2024 Jan 18;11:1332878. doi: 10.3389/fmolb.2024.1332878. eCollection 2024. Front Mol Biosci. 2024. PMID: 38304578 Free PMC article. Review.
-
Linolenic Acid-Derived Oxylipins Inhibit Aflatoxin Biosynthesis in Aspergillus flavus through Activation of Imizoquin Biosynthesis.J Agric Food Chem. 2022 Dec 21;70(50):15928-15944. doi: 10.1021/acs.jafc.2c06230. Epub 2022 Dec 12. J Agric Food Chem. 2022. PMID: 36508213 Free PMC article.
-
Genomic and Phenotypic Trait Variation of the Opportunistic Human Pathogen Aspergillus flavus and Its Close Relatives.Microbiol Spectr. 2022 Dec 21;10(6):e0306922. doi: 10.1128/spectrum.03069-22. Epub 2022 Nov 1. Microbiol Spectr. 2022. PMID: 36318036 Free PMC article.
References
-
- Yang K., Liu Y., Wang S., Wu L., Xie R., Lan H., Fasoyin O.E., Wang Y., Wang S. Cyclase-Associated Protein Cap with Multiple Domains Contributes to Mycotoxin Biosynthesis and Fungal Virulence in Aspergillus flavus. J. Agric. Food Chem. 2019;67:4200–4213. doi: 10.1021/acs.jafc.8b07115. - DOI - PubMed
-
- Chang P.K., Zhang Q., Scharfenstein L., Mack B., Yoshimi A., Miyazawa K., Abe K. Aspergillus flavus GPI-anchored protein-encoding ecm33 has a role in growth, development, aflatoxin biosynthesis, and maize infection. Appl. Microbiol. Biotechnol. 2018;102:5209–5220. doi: 10.1007/s00253-018-9012-7. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

