Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms
- PMID: 33202645
- PMCID: PMC7697839
- DOI: 10.3390/ijms21228594
Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms
Abstract
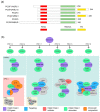
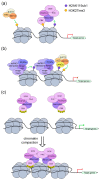
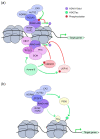
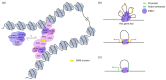
Polycomb group (PcG) proteins function as vital epigenetic regulators in various biological processes, including pluripotency, development, and carcinogenesis. PcG proteins form multicomponent complexes, and two major types of protein complexes have been identified in mammals to date, Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2). The PRC1 complexes are composed in a hierarchical manner in which the catalytic core, RING1A/B, exclusively interacts with one of six Polycomb group RING finger (PCGF) proteins. This association with specific PCGF proteins allows for PRC1 to be subdivided into six distinct groups, each with their own unique modes of action arising from the distinct set of associated proteins. Historically, PRC1 was considered to be a transcription repressor that deposited monoubiquitylation of histone H2A at lysine 119 (H2AK119ub1) and compacted local chromatin. More recently, there is increasing evidence that demonstrates the transcription activation role of PRC1. Moreover, studies on the higher-order chromatin structure have revealed a new function for PRC1 in mediating long-range interactions. This provides a different perspective regarding both the transcription activation and repression characteristics of PRC1. This review summarizes new advancements regarding the composition of mammalian PRC1 and accompanying explanations of how diverse PRC1-associated proteins participate in distinct transcription regulation mechanisms.
Keywords: PRC1; chromatin structure; transcription regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Molecular architecture of polycomb repressive complexes.Biochem Soc Trans. 2017 Feb 8;45(1):193-205. doi: 10.1042/BST20160173. Biochem Soc Trans. 2017. PMID: 28202673 Free PMC article. Review.
-
Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression.Mol Cell. 2020 Feb 20;77(4):840-856.e5. doi: 10.1016/j.molcel.2019.11.021. Epub 2019 Dec 26. Mol Cell. 2020. PMID: 31883952 Free PMC article.
-
Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing.Nat Commun. 2019 Apr 29;10(1):1931. doi: 10.1038/s41467-019-09628-6. Nat Commun. 2019. PMID: 31036804 Free PMC article.
-
Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements.Nucleic Acids Res. 2016 Dec 1;44(21):10132-10149. doi: 10.1093/nar/gkw701. Epub 2016 Aug 23. Nucleic Acids Res. 2016. PMID: 27557709 Free PMC article.
-
Polycomb-mediated histone modifications and gene regulation.Biochem Soc Trans. 2024 Feb 28;52(1):151-161. doi: 10.1042/BST20230336. Biochem Soc Trans. 2024. PMID: 38288743 Review.
Cited by
-
Nuclear PCGF3 inhibits the antiviral immune response by suppressing the interferon-stimulated gene.Cell Death Discov. 2024 Oct 5;10(1):429. doi: 10.1038/s41420-024-02194-x. Cell Death Discov. 2024. PMID: 39368978 Free PMC article.
-
auts2 Features and Expression Are Highly Conserved during Evolution Despite Different Evolutionary Fates Following Whole Genome Duplication.Cells. 2022 Aug 30;11(17):2694. doi: 10.3390/cells11172694. Cells. 2022. PMID: 36078102 Free PMC article.
-
AUTS2 Gene: Keys to Understanding the Pathogenesis of Neurodevelopmental Disorders.Cells. 2021 Dec 21;11(1):11. doi: 10.3390/cells11010011. Cells. 2021. PMID: 35011572 Free PMC article. Review.
-
De Novo Polycomb Recruitment and Repressive Domain Formation.Epigenomes. 2022 Aug 22;6(3):25. doi: 10.3390/epigenomes6030025. Epigenomes. 2022. PMID: 35997371 Free PMC article. Review.
-
Epigenetic Mechanisms of Epidermal Differentiation.Int J Mol Sci. 2022 Apr 28;23(9):4874. doi: 10.3390/ijms23094874. Int J Mol Sci. 2022. PMID: 35563264 Free PMC article. Review.
References
-
- Lewis P.H. New mutants report. Drosoph. Inf. Serv. 1947;21:69.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

