A computational study to disclose potential drugs and vaccine ensemble for COVID-19 conundrum
- PMID: 33199930
- PMCID: PMC7654302
- DOI: 10.1016/j.molliq.2020.114734
A computational study to disclose potential drugs and vaccine ensemble for COVID-19 conundrum
Abstract
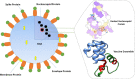
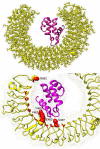
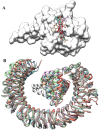
The nucleocapsid (N) protein of SARS-COV-2, a virus responsible for the current COVID-19 pandemic, is considered a potential candidate for the design of new drugs and vaccines. The protein is central to several critical events in virus production, with its highly druggable nature and rich antigenic determinants making it an excellent anti-viral biomolecule. Docking-based virtual screening using the Asinex anti-viral library identified binding of drug molecules at three specific positions: loop 1 region, loop 2 region and β-sheet core pockets, the loop 2 region being the most common binding and stable site for the bulk of the molecules. In parallel, the protein was characterized by vaccine design perspective and harboured three potential B cell-derived T cell epitopes: PINTNSSPD, GVPINTNSS, and DHIGTRNPA. The epitopes are highly antigenic, virulent, non-allergic, non-toxic, bind with good affinity to the highly prevalent DRB*0101 allele and show an average population coverage of 95.04%. A multi-epitope vaccine ensemble which was 83 amino acids long was created. This was highly immunogenic, robust in generating both humoral and cellular immune responses, thermally stable, and had good physicochemical properties that could be easily analyzed in in vivo and in vitro studies. Conformational dynamics of both drug and vaccine ensemble with respect to the receptors are energetically stable, shedding light on favourable conformation and chemical interactions. These facts were validated by subjecting the complexes to relative and absolute binding free energy methods of MMGB/PBSA and WaterSwap. A strong agreement on the system stability was disclosed that supported ligand high affinity potential for the receptors. Collectively, this work sought to provide preliminary experimental data of existing anti-viral drugs as a possible therapy for COVID-19 infections and a new peptide-based vaccine for protection against this pandemic virus.
Keywords: COVID-1; Drugs; Nucleocapsid (N) protein; SARS-COV-2; Vaccine.
© 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors in this study have no conflict of interest.
Figures
Similar articles
-
An in silico study to unveil potential drugs and vaccine chimera for HBV capsid assembly protein: combined molecular docking and dynamics simulation approach.J Mol Model. 2022 Feb 2;28(2):51. doi: 10.1007/s00894-022-05042-w. J Mol Model. 2022. PMID: 35112241
-
Immuno-Informatics Quest against COVID-19/SARS-COV-2: Determining Putative T-Cell Epitopes for Vaccine Prediction.Infect Disord Drug Targets. 2021;21(4):541-552. doi: 10.2174/1871526520666200921154149. Infect Disord Drug Targets. 2021. PMID: 32957905
-
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection.Elife. 2022 Jan 24;11:e73490. doi: 10.7554/eLife.73490. Elife. 2022. PMID: 35072628 Free PMC article.
-
Computational perspectives revealed prospective vaccine candidates from five structural proteins of novel SARS corona virus 2019 (SARS-CoV-2).PeerJ. 2020 Sep 29;8:e9855. doi: 10.7717/peerj.9855. eCollection 2020. PeerJ. 2020. PMID: 33062414 Free PMC article.
-
A reverse vaccinology and immunoinformatics approach for designing a multiepitope vaccine against SARS-CoV-2.Immunogenetics. 2021 Dec;73(6):459-477. doi: 10.1007/s00251-021-01228-3. Epub 2021 Sep 20. Immunogenetics. 2021. PMID: 34542663 Free PMC article.
Cited by
-
Current Treatments for COVID-19: Application of Supercritical Fluids in the Manufacturing of Oral and Pulmonary Formulations.Pharmaceutics. 2022 Nov 4;14(11):2380. doi: 10.3390/pharmaceutics14112380. Pharmaceutics. 2022. PMID: 36365198 Free PMC article. Review.
-
Vaccinomics to Design a Multiepitope Vaccine against Legionella pneumophila.Biomed Res Int. 2022 Sep 17;2022:4975721. doi: 10.1155/2022/4975721. eCollection 2022. Biomed Res Int. 2022. PMID: 36164443 Free PMC article.
-
Comparing the Nucleocapsid Proteins of Human Coronaviruses: Structure, Immunoregulation, Vaccine, and Targeted Drug.Front Mol Biosci. 2022 Apr 29;9:761173. doi: 10.3389/fmolb.2022.761173. eCollection 2022. Front Mol Biosci. 2022. PMID: 35573742 Free PMC article. Review.
-
Design of an Epitope-Based Vaccine Against MERS-CoV.Medicina (Kaunas). 2024 Oct 6;60(10):1632. doi: 10.3390/medicina60101632. Medicina (Kaunas). 2024. PMID: 39459420 Free PMC article.
-
Effect of crystallite size reduction and widening of optical phonon vibration due to AC variation on ZnO/Mg composites in implementation of methylene blue degradation.Photochem Photobiol Sci. 2024 Sep;23(9):1709-1724. doi: 10.1007/s43630-024-00624-4. Epub 2024 Aug 30. Photochem Photobiol Sci. 2024. PMID: 39212858
References
-
- Ahmed T., Noman M., Almatroudi A., Shahid M., Khurshid M., Tariq F., Qamar M.T. ul, Yu R., Li B. 2020. A Novel Coronavirus 2019 Linked with Pneumonia in China: Current Status and Future Prospects.
-
- Chan J.F.-W., Yuan S., Kok K.-H., K.K.-W. To, Chu H., Yang J., Xing F., Liu J., Yip C.C.-Y., Poon R.W.-S., et al. A familial cluster of pneumonia associated with the 2019 Novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
