The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation
- PMID: 33184237
- PMCID: PMC8356967
- DOI: 10.1126/science.abc9359
The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation
Abstract
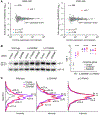
MicroRNAs (miRNAs) associate with Argonaute (AGO) proteins to direct widespread posttranscriptional gene repression. Although association with AGO typically protects miRNAs from nucleases, extensive pairing to some unusual target RNAs can trigger miRNA degradation. We found that this target-directed miRNA degradation (TDMD) required the ZSWIM8 Cullin-RING E3 ubiquitin ligase. This and other findings support a mechanistic model of TDMD in which target-directed proteolysis of AGO by the ubiquitin-proteasome pathway exposes the miRNA for degradation. Moreover, loss-of-function studies indicated that the ZSWIM8 Cullin-RING ligase accelerates degradation of numerous miRNAs in cells of mammals, flies, and nematodes, thereby specifying the half-lives of most short-lived miRNAs. These results elucidate the mechanism of TDMD and expand its inferred role in shaping miRNA levels in bilaterian animals.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


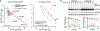

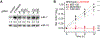

Comment in
-
To Degrade a MicroRNA, Destroy Its Argonaute Protein.Mol Cell. 2021 Jan 21;81(2):223-225. doi: 10.1016/j.molcel.2020.12.043. Mol Cell. 2021. PMID: 33482091
Similar articles
-
A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming.Science. 2020 Dec 18;370(6523):eabc9546. doi: 10.1126/science.abc9546. Epub 2020 Nov 12. Science. 2020. PMID: 33184234 Free PMC article.
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
-
Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins.Genes Dev. 2021 Dec 1;35(23-24):1595-1609. doi: 10.1101/gad.348874.121. Epub 2021 Nov 24. Genes Dev. 2021. PMID: 34819352 Free PMC article.
-
Ago2 protects Drosophila siRNAs and microRNAs from target-directed degradation, even in the absence of 2'-O-methylation.RNA. 2021 Jun;27(6):710-724. doi: 10.1261/rna.078746.121. Epub 2021 Apr 14. RNA. 2021. PMID: 33853897 Free PMC article.
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
Cited by
-
What goes up must come down: off switches for regulatory RNAs.Genes Dev. 2024 Aug 20;38(13-14):597-613. doi: 10.1101/gad.351934.124. Genes Dev. 2024. PMID: 39111824 Free PMC article. Review.
-
Elucidation of the conformational dynamics and assembly of Argonaute-RNA complexes by distinct yet coordinated actions of the supplementary microRNA.Comput Struct Biotechnol J. 2022 Mar 7;20:1352-1365. doi: 10.1016/j.csbj.2022.03.001. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35356544 Free PMC article.
-
The epigenetic regulatory mechanism of PIWI/piRNAs in human cancers.Mol Cancer. 2023 Mar 7;22(1):45. doi: 10.1186/s12943-023-01749-3. Mol Cancer. 2023. PMID: 36882835 Free PMC article. Review.
-
Terminal modification, sequence, length, and PIWI-protein identity determine piRNA stability.Mol Cell. 2021 Dec 2;81(23):4826-4842.e8. doi: 10.1016/j.molcel.2021.09.012. Epub 2021 Oct 8. Mol Cell. 2021. PMID: 34626567 Free PMC article.
-
The Roles of microRNAs in the Cardiovascular System.Int J Mol Sci. 2023 Sep 19;24(18):14277. doi: 10.3390/ijms241814277. Int J Mol Sci. 2023. PMID: 37762578 Free PMC article. Review.
References
-
- Jonas S, Izaurralde E, Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16, 421–433 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

