Genome-Wide CRISPR-Cas9 Screen Reveals the Importance of the Heparan Sulfate Pathway and the Conserved Oligomeric Golgi Complex for Synthetic Double-Stranded RNA Uptake and Sindbis Virus Infection
- PMID: 33177215
- PMCID: PMC7657590
- DOI: 10.1128/mSphere.00914-20
Genome-Wide CRISPR-Cas9 Screen Reveals the Importance of the Heparan Sulfate Pathway and the Conserved Oligomeric Golgi Complex for Synthetic Double-Stranded RNA Uptake and Sindbis Virus Infection
Abstract
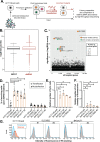
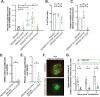
Double-stranded RNA (dsRNA) is the hallmark of many viral infections. dsRNA is produced either by RNA viruses during replication or by DNA viruses upon convergent transcription. Synthetic dsRNA is also able to mimic viral-induced activation of innate immune response and cell death. In this study, we employed a genome-wide CRISPR-Cas9 loss-of-function screen based on cell survival in order to identify genes implicated in the host response to dsRNA. By challenging HCT116 human cells with either synthetic dsRNA or Sindbis virus (SINV), we identified the heparan sulfate (HS) pathway as a crucial factor for dsRNA entry, and we validated SINV dependency on HS. Interestingly, we uncovered a novel role for COG4, a component of the conserved oligomeric Golgi (COG) complex, as a factor involved in cell survival to both dsRNA and SINV in human cells. We showed that COG4 knockout led to a decrease of extracellular HS that specifically affected dsRNA transfection efficiency and reduced viral production, which explains the increased cell survival of these mutants.IMPORTANCE When facing a viral infection, the organism has to put in place a number of defense mechanisms in order to clear the pathogen from the cell. At the early phase of this preparation for fighting against the invader, the innate immune response is triggered by the sensing of danger signals. Among those molecular cues, double-stranded RNA (dsRNA) is a very potent inducer of different reactions at the cellular level that can ultimately lead to cell death. Using a genome-wide screening approach, we set to identify genes involved in dsRNA entry, sensing, and apoptosis induction in human cells. This allowed us to determine that the heparan sulfate pathway and the conserved oligomeric Golgi complex are key determinants allowing entry of both dsRNA and viral nucleic acid leading to cell death.
Keywords: CRISPR-Cas9 screen; complex oligomeric Golgi complex; double-stranded RNA; heparan-sulfate; transfection; virus.
Copyright © 2020 Petitjean et al.
Figures

Similar articles
-
Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses.J Virol. 2015 Sep;89(18):9383-92. doi: 10.1128/JVI.01299-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136565 Free PMC article.
-
Opposing Roles of Double-Stranded RNA Effector Pathways and Viral Defense Proteins Revealed with CRISPR-Cas9 Knockout Cell Lines and Vaccinia Virus Mutants.J Virol. 2016 Aug 12;90(17):7864-79. doi: 10.1128/JVI.00869-16. Print 2016 Sep 1. J Virol. 2016. PMID: 27334583 Free PMC article.
-
A Genome-Wide Haploid Genetic Screen Identifies Heparan Sulfate-Associated Genes and the Macropinocytosis Modulator TMED10 as Factors Supporting Vaccinia Virus Infection.J Virol. 2019 Jun 14;93(13):e02160-18. doi: 10.1128/JVI.02160-18. Print 2019 Jul 1. J Virol. 2019. PMID: 30996093 Free PMC article.
-
Viral infection and host defense.Science. 1974 Dec 27;186(4170):1172-8. doi: 10.1126/science.186.4170.1172. Science. 1974. PMID: 4610750 Review.
-
A Novel Mechanism Underlying the Innate Immune Response Induction upon Viral-Dependent Replication of Host Cell mRNA: A Mistake of +sRNA Viruses' Replicases.Front Cell Infect Microbiol. 2017 Jan 20;7:5. doi: 10.3389/fcimb.2017.00005. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28164038 Free PMC article. Review.
Cited by
-
Genome-wide CRISPR screens and their applications in infectious disease.Front Genome Ed. 2023 Sep 19;5:1243731. doi: 10.3389/fgeed.2023.1243731. eCollection 2023. Front Genome Ed. 2023. PMID: 37794981 Free PMC article. Review.
-
A review of virus host factor discovery using CRISPR screening.mBio. 2024 Nov 13;15(11):e0320523. doi: 10.1128/mbio.03205-23. Epub 2024 Oct 18. mBio. 2024. PMID: 39422472 Free PMC article. Review.
-
Spatiotemporal diversity and regulation of glycosaminoglycans in cell homeostasis and human disease.Am J Physiol Cell Physiol. 2022 May 1;322(5):C849-C864. doi: 10.1152/ajpcell.00085.2022. Epub 2022 Mar 16. Am J Physiol Cell Physiol. 2022. PMID: 35294848 Free PMC article. Review.
-
Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment.Microorganisms. 2021 Jun 7;9(6):1238. doi: 10.3390/microorganisms9061238. Microorganisms. 2021. PMID: 34200288 Free PMC article. Review.
-
Acute COG complex inactivation unveiled its immediate impact on Golgi and illuminated the nature of intra-Golgi recycling vesicles.Traffic. 2023 Feb;24(2):52-75. doi: 10.1111/tra.12876. Epub 2022 Dec 15. Traffic. 2023. PMID: 36468177 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
