Transcriptomic profiles in Parkinson's disease
- PMID: 33148011
- PMCID: PMC7934142
- DOI: 10.1177/1535370220967325
Transcriptomic profiles in Parkinson's disease
Abstract
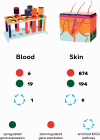
Transcriptomics in Parkinson's disease offers insights into the pathogenesis of Parkinson's disease but obtaining brain tissue has limitations. In order to bypass this issue, we profile and compare differentially expressed genes and enriched pathways (KEGG) in two peripheral tissues (blood and skin) of 12 Parkinson's disease patients and 12 healthy controls using RNA-sequencing technique and validation with RT-qPCR. Furthermore, we compare our results to previous Parkinson's disease post mortem brain tissue and blood results using the robust rank aggregation method. The results show no overlapping differentially expressed genes or enriched pathways in blood vs. skin in our sample sets (25 vs. 1068 differentially expressed genes with an FDR ≤ 0.05; 1 vs. 9 pathways in blood and skin, respectively). A meta-analysis from previous transcriptomic sample sets using either microarrays or RNA-Seq yields a robust rank aggregation list of cortical gene expression changes with 43 differentially expressed genes; a list of substantia nigra changes with 2 differentially expressed genes and a list of blood changes with 1 differentially expressed gene being statistically significant at FDR ≤ 0.05. In cortex 1, KEGG pathway was enriched, four in substantia nigra and two in blood. None of the differentially expressed genes or pathways overlap between these tissues. When comparing our previously published skin transcription analysis, two differentially expressed genes between the cortex robust rank aggregation and skin overlap. In this study, for the first time a meta-analysis is applied on transcriptomic sample sets in Parkinson's disease. Simultaneously, it explores the notion that Parkinson's disease is not just a neuronal tissue disease by exploring peripheral tissues. The comparison of different Parkinson's disease tissues yields surprisingly few significant differentially expressed genes and pathways, suggesting that divergent gene expression profiles in distinct cell lineages, metabolic and possibly iatrogenic effects create too much transcriptomic noise for detecting significant signal. On the other hand, there are signs that point towards Parkinson's disease-specific changes in non-neuronal peripheral tissues in Parkinson's disease, indicating that Parkinson's disease might be a multisystem disorder.
Keywords: Biomarkers; blood; brain; neurodegeneration; skin; transcriptomics.
Conflict of interest statement
Figures


Similar articles
-
Looking beyond the brain to improve the pathogenic understanding of Parkinson's disease: implications of whole transcriptome profiling of Patients' skin.BMC Neurol. 2017 Jan 10;17(1):6. doi: 10.1186/s12883-016-0784-z. BMC Neurol. 2017. PMID: 28068941 Free PMC article.
-
Blood transcriptomics of drug-naïve sporadic Parkinson's disease patients.BMC Genomics. 2015 Oct 28;16:876. doi: 10.1186/s12864-015-2058-3. BMC Genomics. 2015. PMID: 26510930 Free PMC article.
-
Transcriptomic Profiling of Circular RNA in Different Brain Regions of Parkinson's Disease in a Mouse Model.Int J Mol Sci. 2020 Apr 24;21(8):3006. doi: 10.3390/ijms21083006. Int J Mol Sci. 2020. PMID: 32344560 Free PMC article.
-
Expanding the search for genetic biomarkers of Parkinson's disease into the living brain.Neurobiol Dis. 2020 Jul;140:104872. doi: 10.1016/j.nbd.2020.104872. Epub 2020 Apr 14. Neurobiol Dis. 2020. PMID: 32302674 Review.
-
A review of genome-wide transcriptomics studies in Parkinson's disease.Eur J Neurosci. 2018 Jan;47(1):1-16. doi: 10.1111/ejn.13760. Epub 2017 Nov 22. Eur J Neurosci. 2018. PMID: 29068110 Review.
Cited by
-
Longitudinal intronic RNA-Seq analysis of Parkinson's disease patients reveals disease-specific nascent transcription.Exp Biol Med (Maywood). 2022 Jun;247(11):945-957. doi: 10.1177/15353702221081027. Epub 2022 Mar 15. Exp Biol Med (Maywood). 2022. PMID: 35289213 Free PMC article.
-
Network Analysis Performed on Transcriptomes of Parkinson's Disease Patients Reveals Dysfunction in Protein Translation.Int J Mol Sci. 2024 Jan 21;25(2):1299. doi: 10.3390/ijms25021299. Int J Mol Sci. 2024. PMID: 38279299 Free PMC article.
-
Druggable targets for Parkinson's disease: transcriptomics based Mendelian randomization study.Sci Rep. 2024 Oct 28;14(1):25763. doi: 10.1038/s41598-024-77401-x. Sci Rep. 2024. PMID: 39468243 Free PMC article.
-
Expression Quantitative Trait Loci (eQTLs) Associated with Retrotransposons Demonstrate their Modulatory Effect on the Transcriptome.Int J Mol Sci. 2021 Jun 12;22(12):6319. doi: 10.3390/ijms22126319. Int J Mol Sci. 2021. PMID: 34204806 Free PMC article.
-
Paradigm shift required for translational research on the brain.Exp Mol Med. 2024 May;56(5):1043-1054. doi: 10.1038/s12276-024-01218-x. Epub 2024 May 1. Exp Mol Med. 2024. PMID: 38689090 Free PMC article. Review.
References
-
- Hughes AJ, Daniel SE, Lees AJ. Improved accuracy of clinical diagnosis of Lewy body Parkinson’s disease. Neurology 2001; 57:1497–9 - PubMed
-
- Postuma RB, Berg D, Stern M, Poewe W, Olanow CW, Oertel W, Obeso J, Marek K, Litvan I, Lang AE, Halliday G, Goetz CG, Gasser T, Dubois B, Chan P, Bloem BR, Adler CH, Deuschl G. MDS clinical diagnostic criteria for Parkinson’s disease: MDS-PD clinical diagnostic criteria. Mov Disord 2015; 30:1591–601 - PubMed
-
- Verstraeten A, Theuns J, Van Broeckhoven C. Progress in unraveling the genetic etiology of parkinson disease in a genomic era. Trends Genet 2015; 31:140–9 - PubMed
-
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, Schulte C, Keller MF, Arepalli S, Letson C, Edsall C, Stefansson H, Liu X, Pliner H, Lee JH, Cheng R; International Parkinson's Disease Genomics Consortium (IPDGC); Parkinson's Study Group (PSG) Parkinson's Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson's Disease Consortium; Alzheimer Genetic Analysis Group, Ikram MA, Ioannidis JP, Hadjigeorgiou GM, Bis JC, Martinez M, Perlmutter JS, Goate A, Marder K, Fiske B, Sutherland M, Xiromerisiou G, Myers RH, Clark LN, Stefansson K, Hardy JA, Heutink P, Chen H, Wood NW, Houlden H, Payami H, Brice A, Scott WK, Gasser T, Bertram L, Eriksson N, Foroud T, Singleton AB.. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet 2014; 46:989–93 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

