Implications of Splicing Alterations in the Onset and Phenotypic Variability of a Family with Subclinical Manifestation of Peutz-Jeghers Syndrome: Bioinformatic and Molecular Evidence
- PMID: 33147782
- PMCID: PMC7662643
- DOI: 10.3390/ijms21218201
Implications of Splicing Alterations in the Onset and Phenotypic Variability of a Family with Subclinical Manifestation of Peutz-Jeghers Syndrome: Bioinformatic and Molecular Evidence
Abstract
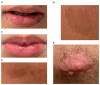
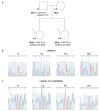
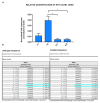
Peutz-Jeghers Syndrome (PJS) is an autosomal dominant pre-cancerous disorder caused in 80-90% of cases by germline mutations in the tumor suppressor gene STK11. We performed a genetic test of the STK11 gene in two Italian young sisters suspected of PJS, since they showed pathognomonic café au lait spots in absence of other symptoms and familiarity. Sequencing of all exons of STK11 gene and other 8 genes, suggested to be involved in hamartomatous syndromes, (PTEN, BMPR1A, SDHB, SDHD, SMAD4, AKT1, ENG, PIK3CA) led to the identification in both the probands of a novel germline silent mutation named c.597 G>A, hitting the last nucleotide of exon 4. Interestingly, genetic testing of the two probands' parents showed that their unaffected father was carrier of this mutation. Moreover, he carried a second intronic substitution named c.465-51 T>C (rs2075606) which was not inherited by his daughters. We also observed that all the family members carrying the c.597 G>A mutation presented an aberrant splice variant of STK11 mRNA lacking exon 4. Furthermore, in silico analysis of c.465-51 T>C substitution showed that it may activate an Enhancer Splicing Element. Finally, qRT-PCR analysis of STK11 expression levels showed a slight downregulation of the wild type allele in the father and a 2-fold downregulation in the probands compared to the unaffected mother. Our results have led the hypothesis that the c.465-51 T>C intronic variant, which segregates with the wild type allele, could increase the splicing effectiveness of STK11 wild-type allele and compensate the side effect of the c.597 G>A splicing mutation, being responsible for the phenotypic variability observed within this family. This finding highlight the importance of RNA analysis in genetic testing, remarking that silent DNA variant can often be splicing variant involved in disease onset and progression. The identification of these variants has a crucial role to ensure an appropriate follow-up and cancer prevention in at-risk individuals.
Keywords: Enhancer Splicing Element; Peutz–Jeghers Syndrome (PJS); café au lait spots; cancer prevention; presymptomatic diagnosis; risk management; splicing variants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Mutation screening at the RNA level of the STK11/LKB1 gene in Peutz-Jeghers syndrome reveals complex splicing abnormalities and a novel mRNA isoform (STK11 c.597(insertion mark)598insIVS4).Hum Mutat. 2001 Nov;18(5):397-410. doi: 10.1002/humu.1211. Hum Mutat. 2001. PMID: 11668633
-
Genetic variation at a splicing branch point in intron 7 of STK11: a rare variant decreasing its expression in a Chinese family with Peutz-Jeghers syndrome.World J Surg Oncol. 2024 Jul 30;22(1):202. doi: 10.1186/s12957-024-03475-6. World J Surg Oncol. 2024. PMID: 39080663 Free PMC article.
-
High prevalence of germline STK11 mutations in Hungarian Peutz-Jeghers Syndrome patients.BMC Med Genet. 2010 Nov 30;11:169. doi: 10.1186/1471-2350-11-169. BMC Med Genet. 2010. PMID: 21118512 Free PMC article.
-
Genetic screening for Peutz-Jeghers syndrome.Expert Rev Mol Diagn. 2003 Jul;3(4):471-9. doi: 10.1586/14737159.3.4.471. Expert Rev Mol Diagn. 2003. PMID: 12877386 Review.
-
[Peutz-Jeghers syndrome].Nihon Rinsho. 2000 Jul;58(7):1400-4. Nihon Rinsho. 2000. PMID: 10921312 Review. Japanese.
Cited by
-
Cancer Risk of Peutz-Jeghers Syndrome and Treatment Experience: A Chinese Medical Center.Clin Colon Rectal Surg. 2023 May 3;36(6):406-414. doi: 10.1055/s-0043-1767704. eCollection 2023 Nov. Clin Colon Rectal Surg. 2023. PMID: 37795464 Free PMC article. Review.
-
Biomarkers in Rare Diseases.Int J Mol Sci. 2021 Jan 12;22(2):673. doi: 10.3390/ijms22020673. Int J Mol Sci. 2021. PMID: 33445477 Free PMC article.
-
Molecular biomarkers, network biomarkers, and dynamic network biomarkers for diagnosis and prediction of rare diseases.Fundam Res. 2022 Aug 9;2(6):894-902. doi: 10.1016/j.fmre.2022.07.011. eCollection 2022 Nov. Fundam Res. 2022. PMID: 38933388 Free PMC article. Review.
References
-
- Sado T., Nakayama Y., Kato S., Homma H., Kusakari M., Hidaka N., Gomi S., Takamizawa S., Kosho T., Saito S., et al. Extremely Young Case of Small Bowel Intussusception Due to Peutz–Jeghers Syndrome with Nonsense Mutation of STK11. Clin. J. Gastroenterol. 2019;12:429–433. doi: 10.1007/s12328-019-00964-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

