Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens
- PMID: 33142918
- PMCID: PMC7672628
- DOI: 10.3390/ijms21218170
Whole-Transcriptome Sequencing-Based Analysis of DAZL and Its Interacting Genes during Germ Cells Specification and Zygotic Genome Activation in Chickens
Abstract
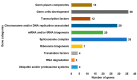
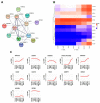
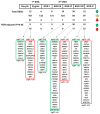
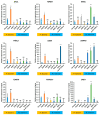
The deleted in azoospermia like (DAZL) is required for germ cells development and maintenance. In chickens, the mRNA and protein of DAZL, a representative maternally inherited germ plasm factor, are detected in the germ plasm of oocyte, zygote, and all stages of the intrauterine embryos. However, it is still insufficient to explain the origin and specification process of chicken germ cells, because the stage at which the zygotic transcription of DAZL occurs and the stage at which the maternal DAZL RNA/protein clears have not yet been fully identified. Moreover, a comprehensive understanding of the expression of DAZL interacting genes during the germ cells specification and development and zygotic genome activation (ZGA) is lacking in chickens. In this study, we identified a set of DAZL interacting genes in chickens using in silico prediction method. Then, we analyzed the whole-transcriptome sequencing (WTS)-based expression of DAZL and its interacting genes in the chicken oocyte, zygote, and Eyal-Giladi and Kochav (EGK) stage embryos (EGK.I to EGK.X). In the results, DAZL transcripts are increased in the zygote (onset of transcription), maintained the increased level until EGK.VI, and decreased from EGK.VIII (possible clearance of maternal RNAs). Among the DAZL interacting genes, most of them are increased either at 1st ZGA or 2nd ZGA, indicating their involvement in germ cells specification and development.
Keywords: DAZL; DAZL interacting genes; germ cells development; intrauterine embryos; zygotic genome activation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Zygotic genome activation in the chicken: a comparative review.Cell Mol Life Sci. 2020 May;77(10):1879-1891. doi: 10.1007/s00018-019-03360-6. Epub 2019 Nov 15. Cell Mol Life Sci. 2020. PMID: 31728579 Free PMC article. Review.
-
DAZL Expression Explains Origin and Central Formation of Primordial Germ Cells in Chickens.Stem Cells Dev. 2016 Jan 1;25(1):68-79. doi: 10.1089/scd.2015.0208. Epub 2015 Oct 27. Stem Cells Dev. 2016. PMID: 26414995
-
The first whole transcriptomic exploration of pre-oviposited early chicken embryos using single and bulked embryonic RNA-sequencing.Gigascience. 2018 Apr 1;7(4):1-9. doi: 10.1093/gigascience/giy030. Gigascience. 2018. PMID: 29659814 Free PMC article.
-
Conserved expression pattern of chicken DAZL in primordial germ cells and germ-line cells.Theriogenology. 2010 Sep 15;74(5):765-76. doi: 10.1016/j.theriogenology.2010.04.001. Epub 2010 May 26. Theriogenology. 2010. PMID: 20537692
-
Zygotic Genome Activation Revisited: Looking Through the Expression and Function of Zscan4.Curr Top Dev Biol. 2016;120:103-24. doi: 10.1016/bs.ctdb.2016.04.004. Epub 2016 May 31. Curr Top Dev Biol. 2016. PMID: 27475850 Review.
Cited by
-
Finer resolution analysis of transcriptional programming during the active migration of chicken primordial germ cells.Comput Struct Biotechnol J. 2022 Oct 26;20:5911-5924. doi: 10.1016/j.csbj.2022.10.034. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36382185 Free PMC article.
-
Dissecting chicken germ cell dynamics by combining a germ cell tracing transgenic chicken model with single-cell RNA sequencing.Comput Struct Biotechnol J. 2022 Apr 2;20:1654-1669. doi: 10.1016/j.csbj.2022.03.040. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35465157 Free PMC article.
-
Fate Decisions of Chicken Primordial Germ Cells (PGCs): Development, Integrity, Sex Determination, and Self-Renewal Mechanisms.Genes (Basel). 2023 Feb 28;14(3):612. doi: 10.3390/genes14030612. Genes (Basel). 2023. PMID: 36980885 Free PMC article. Review.
-
Bioinformatics Applications to Reveal Molecular Mechanisms of Gene Expression Regulation in Model Organisms.Int J Mol Sci. 2021 Nov 5;22(21):11973. doi: 10.3390/ijms222111973. Int J Mol Sci. 2021. PMID: 34769403 Free PMC article.
-
Dynamic maternal synthesis and segregation of the germ plasm organizer, Bucky ball, in chicken oocytes and follicles.Sci Rep. 2024 Nov 12;14(1):27753. doi: 10.1038/s41598-024-78544-7. Sci Rep. 2024. PMID: 39532932 Free PMC article.
References
-
- Hird S.N., Paulsen J.E., Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: Cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

