Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers
- PMID: 33141028
- PMCID: PMC7969425
- DOI: 10.1016/j.pharmthera.2020.107715
Circular RNAs are a novel type of non-coding RNAs in ROS regulation, cardiovascular metabolic inflammations and cancers
Abstract
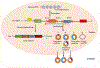
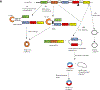
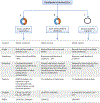
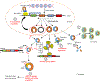
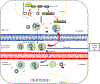
Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNAs characterized by a covalently closed-loop structure generated through a special type of alternative splicing termed back-splicing. Currently, an increasing body of evidence has demonstrated that 1) majority of circRNAs are evolutionarily conserved across species, stable, and resistant to RNase R degradation, and often exhibit cell-specific, and tissue-specific/developmental-stage-specific expression and can be largely independent of the expression levels of the linear host gene-encoded linear RNAs; 2) the biogenesis of circRNAs via back-splicing is different from the canonical splicing of linear RNAs; 3) circRNA biogenesis is regulated by specific cis-acting elements and trans-acting factors; 4) circRNAs regulate biological and pathological processes by sponging miRNAs, binding to RNA-binding protein (RBP), regulators of splicing and transcription, modifiers of parental gene expression, and regulators of protein translation or being translated into peptides in various diseases; 5) circRNAs have been identified for their enrichment and stability in exosomes and detected in body fluids such as human blood, saliva, and cerebrospinal fluids, suggesting that these exo-circRNAs have potential applications as disease biomarkers and novel therapeutic targets; 6) several circRNAs are regulated by oxidative stress and mediate reactive oxygen species (ROS) production as well as promote ROS-induced cellular death, cell apoptosis, and inflammation; 7) circRNAs have also emerged as important regulators in atherosclerotic cardiovascular disease, metabolic disease, and cancers; 8) the potential mechanisms of several circRNAs have been described in diseases, hinting at their potential applications as novel therapeutic targets. In this highlight, we summarized the current understandings of the biogenesis and functions of circRNAs and their roles in ROS regulation and vascular inflammation-associated with cardiovascular and metabolic disease. (Word count: 272).
Keywords: Circular RNAs (circRNAs); Exosomes; Reactive oxygen species (ROS); Vascular inflammation.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Review on circular RNAs and new insights into their roles in cancer.Comput Struct Biotechnol J. 2021 Jan 22;19:910-928. doi: 10.1016/j.csbj.2021.01.018. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33598105 Free PMC article. Review.
-
Circular RNA: A new star of noncoding RNAs.Cancer Lett. 2015 Sep 1;365(2):141-8. doi: 10.1016/j.canlet.2015.06.003. Epub 2015 Jun 5. Cancer Lett. 2015. PMID: 26052092 Review.
-
CircRNAs and their regulatory roles in cancers.Mol Med. 2021 Aug 26;27(1):94. doi: 10.1186/s10020-021-00359-3. Mol Med. 2021. PMID: 34445958 Free PMC article. Review.
-
Circular RNAs: An emerging type of RNA in cancer.Int J Immunopathol Pharmacol. 2017 Mar;30(1):1-6. doi: 10.1177/0394632016686985. Epub 2017 Jan 30. Int J Immunopathol Pharmacol. 2017. PMID: 28134598 Free PMC article.
-
Decrypting the circular RNAs does a favor for us: Understanding, diagnosing and treating diabetes mellitus and its complications.Biomed Pharmacother. 2023 Dec;168:115744. doi: 10.1016/j.biopha.2023.115744. Epub 2023 Oct 18. Biomed Pharmacother. 2023. PMID: 37862970 Review.
Cited by
-
Circular RNAs in lung cancer: implications for preventing therapeutic resistance.EBioMedicine. 2024 Sep;107:105309. doi: 10.1016/j.ebiom.2024.105309. Epub 2024 Aug 26. EBioMedicine. 2024. PMID: 39191172 Free PMC article. Review.
-
Identification of Cardiac CircRNAs in Mice With CVB3-Induced Myocarditis.Front Cell Dev Biol. 2022 Feb 7;10:760509. doi: 10.3389/fcell.2022.760509. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35198554 Free PMC article.
-
Oxidative Stress in the Pathogenesis of Aorta Diseases as a Source of Potential Biomarkers and Therapeutic Targets, with a Particular Focus on Ascending Aorta Aneurysms.Antioxidants (Basel). 2022 Jan 18;11(2):182. doi: 10.3390/antiox11020182. Antioxidants (Basel). 2022. PMID: 35204065 Free PMC article. Review.
-
A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6.Clin Transl Med. 2021 Oct;11(10):e613. doi: 10.1002/ctm2.613. Clin Transl Med. 2021. PMID: 34709743 Free PMC article.
-
CircRNA7632 down-regulation alleviates endothelial cell dysfunction in Kawasaki disease via regulating IL-33 expression.Cell Stress Chaperones. 2023 Jul;28(4):363-374. doi: 10.1007/s12192-023-01333-0. Epub 2023 May 11. Cell Stress Chaperones. 2023. PMID: 37166618 Free PMC article.
References
-
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. (2014). Circrna biogenesis competes with pre-mrna splicing. Molecular Cell 56, 55–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

