Essential role of the linear ubiquitin chain assembly complex and TAK1 kinase in A20 mutant Hodgkin lymphoma
- PMID: 33139544
- PMCID: PMC7682429
- DOI: 10.1073/pnas.2014470117
Essential role of the linear ubiquitin chain assembly complex and TAK1 kinase in A20 mutant Hodgkin lymphoma
Abstract
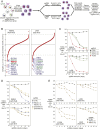
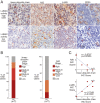
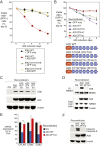
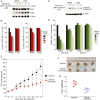
More than 70% of Epstein-Barr virus (EBV)-negative Hodgkin lymphoma (HL) cases display inactivation of TNFAIP3 (A20), a ubiquitin-editing protein that regulates nonproteolytic protein ubiquitination, indicating the significance of protein ubiquitination in HL pathogenesis. However, the precise mechanistic roles of A20 and the ubiquitination system remain largely unknown in this disease. Here, we performed high-throughput CRISPR screening using a ubiquitin regulator-focused single-guide RNA library in HL lines carrying either wild-type or mutant A20. Our CRISPR screening highlights the essential oncogenic role of the linear ubiquitin chain assembly complex (LUBAC) in HL lines, which overlaps with A20 inactivation status. Mechanistically, LUBAC promotes IKK/NF-κB activity and NEMO linear ubiquitination in A20 mutant HL cells, which is required for prosurvival genes and immunosuppressive molecule expression. As a tumor suppressor, A20 directly inhibits IKK activation and HL cell survival via its C-terminal linear-ubiquitin binding ZF7. Clinically, LUBAC activity is consistently elevated in most primary HL cases, and this is correlated with high NF-κB activity and low A20 expression. To further understand the complete mechanism of NF-κB activation in A20 mutant HL, we performed a specifically designed CD83-based NF-κB CRISPR screen which led us to identify TAK1 kinase as a major mediator for NF-κB activation in cells dependent on LUBAC, where the LUBAC-A20 axis regulates TAK1 and IKK complex formation. Finally, TAK1 inhibitor Takinib shows promising activity against HL in vitro and in a xenograft mouse model. Altogether, these findings provide strong support that targeting LUBAC or TAK1 could be attractive therapeutic strategies in A20 mutant HL.
Keywords: Hodgkin lymphoma; NF-kB; signaling transduction; ubiquitination.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7.EMBO J. 2012 Oct 3;31(19):3845-55. doi: 10.1038/emboj.2012.240. Epub 2012 Aug 28. EMBO J. 2012. PMID: 23032186 Free PMC article.
-
Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation.EMBO J. 2012 Oct 3;31(19):3856-70. doi: 10.1038/emboj.2012.241. Epub 2012 Aug 28. EMBO J. 2012. PMID: 23032187 Free PMC article.
-
Porcine Reproductive and Respiratory Syndrome Virus nsp1α Inhibits NF-κB Activation by Targeting the Linear Ubiquitin Chain Assembly Complex.J Virol. 2017 Jan 18;91(3):e01911-16. doi: 10.1128/JVI.01911-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881655 Free PMC article.
-
A20-mediated negative regulation of canonical NF-κB signaling pathway.Immunol Res. 2013 Dec;57(1-3):166-71. doi: 10.1007/s12026-013-8463-2. Immunol Res. 2013. PMID: 24242761 Review.
-
Linear ubiquitination-mediated NF-κB regulation and its related disorders.J Biochem. 2013 Oct;154(4):313-23. doi: 10.1093/jb/mvt079. Epub 2013 Aug 21. J Biochem. 2013. PMID: 23969028 Review.
Cited by
-
Ubiquitination and deubiquitination in cancer: from mechanisms to novel therapeutic approaches.Mol Cancer. 2024 Jul 25;23(1):148. doi: 10.1186/s12943-024-02046-3. Mol Cancer. 2024. PMID: 39048965 Free PMC article. Review.
-
Identification and molecular analysis of RNF31 Q622H germline polymorphism.Oncol Lett. 2022 Sep 21;24(5):394. doi: 10.3892/ol.2022.13514. eCollection 2022 Nov. Oncol Lett. 2022. PMID: 36276481 Free PMC article.
-
Enhanced pathogenicity by up-regulation of A20 after avian leukemia subgroup a virus infection.Front Vet Sci. 2022 Nov 14;9:1031480. doi: 10.3389/fvets.2022.1031480. eCollection 2022. Front Vet Sci. 2022. PMID: 36452148 Free PMC article.
-
Analysis and therapeutic targeting of the IL-1R pathway in anaplastic large cell lymphoma.Blood. 2023 Oct 12;142(15):1297-1311. doi: 10.1182/blood.2022019166. Blood. 2023. PMID: 37339580 Free PMC article.
-
High-Throughput CRISPR Screening in Hematological Neoplasms.Cancers (Basel). 2022 Jul 25;14(15):3612. doi: 10.3390/cancers14153612. Cancers (Basel). 2022. PMID: 35892871 Free PMC article. Review.
References
-
- Bräuninger A., et al. , Molecular biology of Hodgkin’s and Reed/Sternberg cells in Hodgkin’s lymphoma. Int. J. Cancer 118, 1853–1861 (2006). - PubMed
-
- Canellos G. P., et al. , Chemotherapy of advanced Hodgkin’s disease with MOPP, ABVD, or MOPP alternating with ABVD. N. Engl. J. Med. 327, 1478–1484 (1992). - PubMed
-
- Santoro A., et al. , Long-term results of combined chemotherapy-radiotherapy approach in Hodgkin’s disease: Superiority of ABVD plus radiotherapy versus MOPP plus radiotherapy. J. Clin. Oncol. 5, 27–37 (1987). - PubMed
-
- Weniger M. A., Küppers R., NF-κB deregulation in Hodgkin lymphoma. Semin. Cancer Biol. 39, 32–39 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

