Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes
- PMID: 33137199
- PMCID: PMC7672446
- DOI: 10.1093/nar/gkaa935
Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes
Abstract
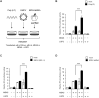
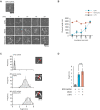
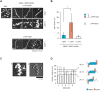
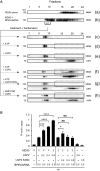
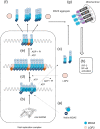
Cytoplasmic RIG-I-like receptor (RLR) proteins in mammalian cells recognize viral RNA and initiate an antiviral response that results in IFN-β induction. Melanoma differentiation-associated protein 5 (MDA5) forms fibers along viral dsRNA and propagates an antiviral response via a signaling domain, the tandem CARD. The most enigmatic RLR, laboratory of genetics and physiology (LGP2), lacks the signaling domain but functions in viral sensing through cooperation with MDA5. However, it remains unclear how LGP2 coordinates fiber formation and subsequent MDA5 activation. We utilized biochemical and biophysical approaches to observe fiber formation and the conformation of MDA5. LGP2 facilitated MDA5 fiber assembly. LGP2 was incorporated into the fibers with an average inter-molecular distance of 32 nm, suggesting the formation of hetero-oligomers with MDA5. Furthermore, limited protease digestion revealed that LGP2 induces significant conformational changes on MDA5, promoting exposure of its CARDs. Although the fibers were efficiently dissociated by ATP hydrolysis, MDA5 maintained its active conformation to participate in downstream signaling. Our study demonstrated the coordinated actions of LGP2 and MDA5, where LGP2 acts as an MDA5 nucleator and requisite partner in the conversion of MDA5 to an active conformation. We revealed a mechanistic basis for LGP2-mediated regulation of MDA5 antiviral innate immune responses.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Quantitative microspectroscopic imaging reveals viral and cellular RNA helicase interactions in live cells.J Biol Chem. 2017 Jul 7;292(27):11165-11177. doi: 10.1074/jbc.M117.777045. Epub 2017 May 8. J Biol Chem. 2017. PMID: 28483922 Free PMC article.
-
LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses.Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1512-7. doi: 10.1073/pnas.0912986107. Epub 2010 Jan 8. Proc Natl Acad Sci U S A. 2010. PMID: 20080593 Free PMC article.
-
LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling.Cytokine. 2015 Aug;74(2):198-206. doi: 10.1016/j.cyto.2015.02.010. Epub 2015 Mar 18. Cytokine. 2015. PMID: 25794939 Free PMC article. Review.
-
Contrasting functions of ATP hydrolysis by MDA5 and LGP2 in viral RNA sensing.J Biol Chem. 2024 Mar;300(3):105711. doi: 10.1016/j.jbc.2024.105711. Epub 2024 Feb 1. J Biol Chem. 2024. PMID: 38309507 Free PMC article.
-
MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction.J Virol. 2014 Aug;88(15):8194-200. doi: 10.1128/JVI.00640-14. Epub 2014 May 21. J Virol. 2014. PMID: 24850739 Free PMC article. Review.
Cited by
-
The molecular mechanism of RIG-I activation and signaling.Immunol Rev. 2021 Nov;304(1):154-168. doi: 10.1111/imr.13022. Epub 2021 Sep 12. Immunol Rev. 2021. PMID: 34514601 Free PMC article. Review.
-
Unraveling blunt-end RNA binding and ATPase-driven translocation activities of the RIG-I family helicase LGP2.Nucleic Acids Res. 2024 Jan 11;52(1):355-369. doi: 10.1093/nar/gkad1106. Nucleic Acids Res. 2024. PMID: 38015453 Free PMC article.
-
The role of TBK1 in cancer pathogenesis and anticancer immunity.J Exp Clin Cancer Res. 2022 Apr 9;41(1):135. doi: 10.1186/s13046-022-02352-y. J Exp Clin Cancer Res. 2022. PMID: 35395857 Free PMC article. Review.
-
RNA sensing via the RIG-I-like receptor LGP2 is essential for the induction of a type I IFN response in ADAR1 deficiency.EMBO J. 2022 Mar 15;41(6):e109760. doi: 10.15252/embj.2021109760. Epub 2022 Feb 14. EMBO J. 2022. PMID: 35156720 Free PMC article.
-
Cellular functions of eukaryotic RNA helicases and their links to human diseases.Nat Rev Mol Cell Biol. 2023 Oct;24(10):749-769. doi: 10.1038/s41580-023-00628-5. Epub 2023 Jul 20. Nat Rev Mol Cell Biol. 2023. PMID: 37474727 Review.
References
-
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T.. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004; 5:730–737. - PubMed
-
- Onomoto K., Onoguchi K., Takahasi K., Fujita T.. Type I interferon production induced by RIG-I-like receptors. J. Interf. Cytokine Res. 2010; 30:875–881. - PubMed
-
- Seth R.B., Sun L., Ea C.K., Chen Z.J.. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005; 122:669–682. - PubMed

