RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3
- PMID: 33129345
- PMCID: PMC7603783
- DOI: 10.1186/s40478-020-01060-y
RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3
Abstract
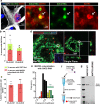
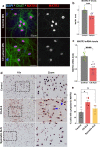
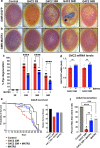
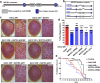
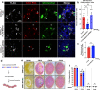
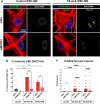
The most common genetic cause of amyotrophic lateral sclerosis (ALS) is a GGGGCC (G4C2) hexanucleotide repeat expansions in first intron of the C9orf72 gene. The accumulation of repetitive RNA sequences can mediate toxicity potentially through the formation of intranuclear RNA foci that sequester key RNA-binding proteins (RBPs), and non-ATG mediated translation into toxic dipeptide protein repeats. However, the contribution of RBP sequestration to the mechanisms underlying RNA-mediated toxicity remain unknown. Here we show that the ALS-associated RNA-binding protein, Matrin-3 (MATR3), colocalizes with G4C2 RNA foci in patient tissues as well as iPSC-derived motor neurons harboring the C9orf72 mutation. Hyperexpansion of C9 repeats perturbed subcellular distribution and levels of endogenous MATR3 in C9-ALS patient-derived motor neurons. Interestingly, we observed that ectopic expression of human MATR3 strongly mitigates G4C2-mediated neurodegeneration in vivo. MATR3-mediated suppression of C9 toxicity was dependent on the RNA-binding domain of MATR3. Importantly, we found that expression of MATR3 reduced the levels of RAN-translation products in mammalian cells in an RNA-dependent manner. Finally, we have shown that knocking down endogenous MATR3 in C9-ALS patient-derived iPSC neurons decreased the presence of G4C2 RNA foci in the nucleus. Overall, these studies suggest that MATR3 genetically modifies the neuropathological and the pathobiology of C9orf72 ALS through modulating the RNA foci and RAN translation.
Conflict of interest statement
The authors declare that they have no competing interests
Figures
Similar articles
-
Modelling C9ORF72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis and frontotemporal dementia.Acta Neuropathol. 2014 Mar;127(3):377-89. doi: 10.1007/s00401-013-1235-1. Epub 2013 Dec 24. Acta Neuropathol. 2014. PMID: 24366528 Review.
-
The exocyst subunit EXOC2 regulates the toxicity of expanded GGGGCC repeats in C9ORF72-ALS/FTD.Cell Rep. 2024 Jul 23;43(7):114375. doi: 10.1016/j.celrep.2024.114375. Epub 2024 Jun 26. Cell Rep. 2024. PMID: 38935506 Free PMC article.
-
Synaptic dysfunction and altered excitability in C9ORF72 ALS/FTD.Brain Res. 2018 Aug 15;1693(Pt A):98-108. doi: 10.1016/j.brainres.2018.02.011. Epub 2018 Feb 14. Brain Res. 2018. PMID: 29453960 Free PMC article. Review.
-
Disruption of nuclear speckle integrity dysregulates RNA splicing in C9ORF72-FTD/ALS.Neuron. 2024 Oct 23;112(20):3434-3451.e11. doi: 10.1016/j.neuron.2024.07.025. Epub 2024 Aug 23. Neuron. 2024. PMID: 39181135
-
Insights into C9ORF72-Related ALS/FTD from Drosophila and iPSC Models.Trends Neurosci. 2018 Jul;41(7):457-469. doi: 10.1016/j.tins.2018.04.002. Epub 2018 May 2. Trends Neurosci. 2018. PMID: 29729808 Free PMC article. Review.
Cited by
-
Toward understanding the role of genomic repeat elements in neurodegenerative diseases.Neural Regen Res. 2025 Mar 1;20(3):646-659. doi: 10.4103/NRR.NRR-D-23-01568. Epub 2024 Apr 16. Neural Regen Res. 2025. PMID: 38886931 Free PMC article.
-
The role of Matrin-3 in physiology and its dysregulation in disease.Biochem Soc Trans. 2024 Jun 26;52(3):961-972. doi: 10.1042/BST20220585. Biochem Soc Trans. 2024. PMID: 38813817 Free PMC article. Review.
-
Phase separation and pathologic transitions of RNP condensates in neurons: implications for amyotrophic lateral sclerosis, frontotemporal dementia and other neurodegenerative disorders.Front Mol Neurosci. 2023 Sep 1;16:1242925. doi: 10.3389/fnmol.2023.1242925. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37720552 Free PMC article. Review.
-
C9orf72 poly(PR) mediated neurodegeneration is associated with nucleolar stress.iScience. 2023 Jul 28;26(9):107505. doi: 10.1016/j.isci.2023.107505. eCollection 2023 Sep 15. iScience. 2023. PMID: 37664610 Free PMC article.
-
SMN regulates GEMIN5 expression and acts as a modifier of GEMIN5-mediated neurodegeneration.Acta Neuropathol. 2023 Sep;146(3):477-498. doi: 10.1007/s00401-023-02607-8. Epub 2023 Jun 27. Acta Neuropathol. 2023. PMID: 37369805 Free PMC article.
References
-
- Brown RH, Al-Chalabi A (2017) Amyotrophic lateral sclerosis. 10.1056/NEJMra1603471. Epub ahead of print 12 July 2017
Publication types
MeSH terms
Substances
Grants and funding
- R21 NS094921/NS/NINDS NIH HHS/United States
- R21 NS101661/NS/NINDS NIH HHS/United States
- R21 NS100055/NS/NINDS NIH HHS/United States
- F31 NS117084/NS/NINDS NIH HHS/United States
- T32 NS086749/NS/NINDS NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- P30 AG066468/AG/NIA NIH HHS/United States
- R21 NS111768/NS/NINDS NIH HHS/United States
- R01 NS104219/NS/NINDS NIH HHS/United States
- R21 AG064940/AG/NIA NIH HHS/United States
- R01 NS081303/NS/NINDS NIH HHS/United States
- T32 NS041234/NS/NINDS NIH HHS/United States
- R21 NS107761/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

