Update of P2X receptor properties and their pharmacology: IUPHAR Review 30
- PMID: 33125712
- PMCID: PMC8199792
- DOI: 10.1111/bph.15299
Update of P2X receptor properties and their pharmacology: IUPHAR Review 30
Abstract
The known seven mammalian receptor subunits (P2X1-7) form cationic channels gated by ATP. Three subunits compose a receptor channel. Each subunit is a polypeptide consisting of two transmembrane regions (TM1 and TM2), intracellular N- and C-termini, and a bulky extracellular loop. Crystallization allowed the identification of the 3D structure and gating cycle of P2X receptors. The agonist-binding pocket is located at the intersection of two neighbouring subunits. In addition to the mammalian P2X receptors, their primitive ligand-gated counterparts with little structural similarity have also been cloned. Selective agonists for P2X receptor subtypes are not available, but medicinal chemistry supplied a range of subtype-selective antagonists, as well as positive and negative allosteric modulators. Knockout mice and selective antagonists helped to identify pathological functions due to defective P2X receptors, such as male infertility (P2X1), hearing loss (P2X2), pain/cough (P2X3), neuropathic pain (P2X4), inflammatory bone loss (P2X5), and faulty immune reactions (P2X7).
Keywords: (patho)physiological functions; P2X receptors; agonists; antagonists; extracellular ATP; knockout mice; ligand-gated cationic channels.
© 2020 The Authors. British Journal of Pharmacology published by John Wiley & Sons Ltd on behalf of British Pharmacological Society.
Conflict of interest statement
CONFLICT OF INTEREST
F.D.V. is a member of the Scientific Advisory Board of Biosceptre Ltd, a UK-based biotech company involved in the development of P2X7-targeted therapeutic antibodies. The other authors declare no competing interest.
Figures
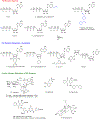


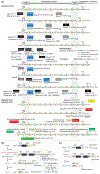
Similar articles
-
Proteomics to identify proteins interacting with P2X2 ligand-gated cation channels.J Vis Exp. 2009 May 18;(27):1178. doi: 10.3791/1178. J Vis Exp. 2009. PMID: 19455095 Free PMC article.
-
P2X receptor chimeras highlight roles of the amino terminus to partial agonist efficacy, the carboxyl terminus to recovery from desensitization, and independent regulation of channel transitions.J Biol Chem. 2013 Jul 19;288(29):21412-21421. doi: 10.1074/jbc.M113.464651. Epub 2013 Jun 5. J Biol Chem. 2013. PMID: 23740251 Free PMC article.
-
Characterization of cultured dorsal root ganglion neuron P2X receptors.Eur J Neurosci. 1999 Jan;11(1):149-54. doi: 10.1046/j.1460-9568.1999.00426.x. Eur J Neurosci. 1999. PMID: 9987019
-
Pharmacology of cloned P2X receptors.Annu Rev Pharmacol Toxicol. 2000;40:563-80. doi: 10.1146/annurev.pharmtox.40.1.563. Annu Rev Pharmacol Toxicol. 2000. PMID: 10836147 Review.
-
Medicinal chemistry of P2X receptors: allosteric modulators.Curr Med Chem. 2015;22(7):929-41. doi: 10.2174/0929867322666141210155610. Curr Med Chem. 2015. PMID: 25524251 Review.
Cited by
-
Purinergic P2 Receptors: Novel Mediators of Mechanotransduction.Front Pharmacol. 2021 May 7;12:671809. doi: 10.3389/fphar.2021.671809. eCollection 2021. Front Pharmacol. 2021. PMID: 34025431 Free PMC article. Review.
-
High-affinity agonism at the P2X7 receptor is mediated by three residues outside the orthosteric pocket.Nat Commun. 2024 Aug 6;15(1):6662. doi: 10.1038/s41467-024-50771-6. Nat Commun. 2024. PMID: 39107314 Free PMC article.
-
Guanine inhibits the growth of human glioma and melanoma cell lines by interacting with GPR23.Front Pharmacol. 2022 Sep 19;13:970891. doi: 10.3389/fphar.2022.970891. eCollection 2022. Front Pharmacol. 2022. PMID: 36199684 Free PMC article.
-
Purinergic signaling: A gatekeeper of blood-brain barrier permeation.Front Pharmacol. 2023 Feb 7;14:1112758. doi: 10.3389/fphar.2023.1112758. eCollection 2023. Front Pharmacol. 2023. PMID: 36825149 Free PMC article. Review.
-
Molecular Pharmacology of P2X Receptors: Exploring Druggable Domains Revealed by Structural Biology.Front Pharmacol. 2022 Jun 17;13:925880. doi: 10.3389/fphar.2022.925880. eCollection 2022. Front Pharmacol. 2022. PMID: 35784697 Free PMC article. Review.
References
-
- Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, … Weisman GA (2006). International Union of Pharmacology LVIII: Update on the P2Y G protein-coupled nucleotide receptors: From molecular mechanisms and pathophysiology to therapy. Pharmacological Reviews, 58, 281–341. 10.1124/pr.58.3.3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 335447717-SFB 1328/Deutsche Forschungsgemeinschaft
- 8YTNWC/Ministry of University and Research
- 13025/Italian Association for Cancer Research
- G20190236012/State Administration of Foreign Experts Affairs
- CZYHW1901/Chengdu University of Traditional Chinese Medicine
- IG 13025/Associazione Italiana per la Ricerca sul Cancro
- Università degli Studi di Ferrara
- 766124/European Union's Horizon 2020
- Z01 DK031127/ImNIH/Intramural NIH HHS/United States
- IG 22883/Associazione Italiana per la Ricerca sul Cancro
- Horizon 2020 Framework Programme
- IG 18581/Associazione Italiana per la Ricerca sul Cancro
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

