Cell Signaling Coordinates Global PRC2 Recruitment and Developmental Gene Expression in Murine Embryonic Stem Cells
- PMID: 33103084
- PMCID: PMC7578752
- DOI: 10.1016/j.isci.2020.101646
Cell Signaling Coordinates Global PRC2 Recruitment and Developmental Gene Expression in Murine Embryonic Stem Cells
Abstract
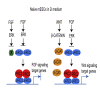
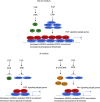
The recruitment of Polycomb repressive complex 2 (PRC2) to gene promoters is critical for its function in repressing gene expression in murine embryonic stem cells (mESCs). However, previous studies have demonstrated that although the expression of early lineage-specific genes is largely repressed, the genome-wide PRC2 occupancy is unexpectedly reduced in naive mESCs. In this study, we provide evidence that fibroblast growth factor/extracellular signal-regulated kinase signaling determines the global PRC2 occupancy through regulating the expression of PRC2-recruiting factor JARID2 in naive mESCs. At the transcriptional level, the de-repression of bivalent genes is predominantly determined by the presence of cell signaling-associated transcription factors but not the status of PRC2 occupancy at gene promoters. Hence, this study not only reveals a key molecular mechanism by which cell signaling regulates the PRC2 occupancy in mESCs but also elucidates the functional roles of transcription factors and Polycomb-mediated epigenetic mechanisms in transcriptional regulation.
Keywords: Cell Biology; Developmental Biology; Molecular Biology; Stem Cells Research.
© 2020 The Author(s).
Conflict of interest statement
Authors declare no competing interests.
Figures

Similar articles
-
Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells.Cell. 2009 Dec 24;139(7):1290-302. doi: 10.1016/j.cell.2009.12.002. Cell. 2009. PMID: 20064375 Free PMC article.
-
JARID2 and the PRC2 complex regulate skeletal muscle differentiation through regulation of canonical Wnt signaling.Epigenetics Chromatin. 2018 Aug 17;11(1):46. doi: 10.1186/s13072-018-0217-x. Epigenetics Chromatin. 2018. PMID: 30119689 Free PMC article.
-
The long noncoding RNA Gm15055 represses Hoxa gene expression by recruiting PRC2 to the gene cluster.Nucleic Acids Res. 2016 Apr 7;44(6):2613-27. doi: 10.1093/nar/gkv1315. Epub 2015 Nov 28. Nucleic Acids Res. 2016. PMID: 26615201 Free PMC article.
-
The Complexity of PRC2 Subcomplexes.Trends Cell Biol. 2019 Aug;29(8):660-671. doi: 10.1016/j.tcb.2019.05.004. Epub 2019 Jun 6. Trends Cell Biol. 2019. PMID: 31178244 Review.
-
Role of PRC2-associated factors in stem cells and disease.FEBS J. 2015 May;282(9):1723-35. doi: 10.1111/febs.13083. Epub 2014 Oct 25. FEBS J. 2015. PMID: 25271128 Review.
Cited by
-
Impaired KDM2B-mediated PRC1 recruitment to chromatin causes defective neural stem cell self-renewal and ASD/ID-like behaviors.iScience. 2022 Jan 7;25(2):103742. doi: 10.1016/j.isci.2022.103742. eCollection 2022 Feb 18. iScience. 2022. PMID: 35128353 Free PMC article.
-
Dynamical modeling of the H3K27 epigenetic landscape in mouse embryonic stem cells.PLoS Comput Biol. 2022 Sep 2;18(9):e1010450. doi: 10.1371/journal.pcbi.1010450. eCollection 2022 Sep. PLoS Comput Biol. 2022. PMID: 36054209 Free PMC article.
-
C3G Regulates STAT3, ERK, Adhesion Signaling, and Is Essential for Differentiation of Embryonic Stem Cells.Stem Cell Rev Rep. 2021 Aug;17(4):1465-1477. doi: 10.1007/s12015-021-10136-8. Epub 2021 Feb 23. Stem Cell Rev Rep. 2021. PMID: 33624208 Free PMC article.
-
Histone H3K36me2-Specific Methyltransferase ASH1L Promotes MLL-AF9-Induced Leukemogenesis.Front Oncol. 2021 Oct 8;11:754093. doi: 10.3389/fonc.2021.754093. eCollection 2021. Front Oncol. 2021. PMID: 34692539 Free PMC article.
-
H3K27me3-mediated epigenetic regulation in pluripotency maintenance and lineage differentiation.Cell Insight. 2024 Jun 27;3(4):100180. doi: 10.1016/j.cellin.2024.100180. eCollection 2024 Aug. Cell Insight. 2024. PMID: 39072246 Free PMC article. Review.
References
-
- Aloia L., Di Stefano B., Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. - PubMed
-
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

