A Department of Defense Laboratory Consortium Approach to Next Generation Sequencing and Bioinformatics Training for Infectious Disease Surveillance in Kenya
- PMID: 33101395
- PMCID: PMC7546821
- DOI: 10.3389/fgene.2020.577563
A Department of Defense Laboratory Consortium Approach to Next Generation Sequencing and Bioinformatics Training for Infectious Disease Surveillance in Kenya
Abstract
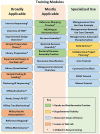
Epidemics of emerging and re-emerging infectious diseases are a danger to civilian and military populations worldwide. Health security and mitigation of infectious disease threats is a priority of the United States Government and the Department of Defense (DoD). Next generation sequencing (NGS) and Bioinformatics (BI) enhances traditional biosurveillance by providing additional data to understand transmission, identify resistance and virulence factors, make predictions, and update risk assessments. As more and more laboratories adopt NGS and BI technologies they encounter challenges in building local capacity. In addition to choosing the right sequencing platform and approach, considerations must also be made for the complexity of bioinformatics analyses, data storage, as well as personnel and computational requirements. To address these needs, a comprehensive training program was developed covering wet lab and bioinformatics approaches to NGS. The program is meant to be modular and adaptive to meet both common and individualized needs of medical research and public health laboratories across the DoD. The training program was first deployed internationally to the Basic Science Laboratory of the US Army Medical Research Directorate-Africa in Kisumu, Kenya, which is an overseas Lab of the Walter Reed Army Institute of Research (WRAIR). A week-long workshop with intensive focus on targeted sequencing and the bioinformatics of genome assembly (n = 24 participants) was held. Post-workshop self-assessment (completed by 21 participants) noted significant median gains in knowledge domains related to NGS targeted sequencing, bioinformatics for genome assembly, and sequence quality assessment. The participants also reported that the information on study design, sample preparation, sequencing quality control, data quality assessment, reporting, and basic and advanced bioinformatics analysis were the most useful information presented in the training. While longer-term evaluations are planned, the training resulted in significant short-term improvement of a laboratory's self-reported wet lab and bioinformatics capabilities. This framework can be used for future DoD laboratory development in the area of NGS and BI for infectious disease surveillance, ultimately enhancing this global DoD capability.
Keywords: DoD; NGS; bioinformatics; infectious disease; workshop.
Copyright © 2020 Maljkovic Berry, Rutvisuttinunt, Voegtly, Prieto, Pollett, Cer, Kugelman, Bishop-Lilly, Morton, Waitumbi and Jarman.
Figures

Similar articles
-
Next-Generation Sequencing and Bioinformatics Consortium Approach to Genomic Surveillance.Emerg Infect Dis. 2024 Oct;30(14):13-18. doi: 10.3201/eid3014.240306. Emerg Infect Dis. 2024. PMID: 39530777 Free PMC article.
-
Next Generation Sequencing and Bioinformatics Methodologies for Infectious Disease Research and Public Health: Approaches, Applications, and Considerations for Development of Laboratory Capacity.J Infect Dis. 2020 Mar 28;221(Suppl 3):S292-S307. doi: 10.1093/infdis/jiz286. J Infect Dis. 2020. PMID: 31612214 Review.
-
Emerging Infectious Diseases and Antimicrobial Resistance (EIDAR).Mil Med. 2019 Nov 1;184(Suppl 2):59-65. doi: 10.1093/milmed/usz081. Mil Med. 2019. PMID: 31004432 Free PMC article.
-
Biomedical Response to Neisseria gonorrhoeae and Other Sexually Transmitted Infections in the US Military.Mil Med. 2019 Nov 1;184(Suppl 2):51-58. doi: 10.1093/milmed/usy431. Mil Med. 2019. PMID: 31778198
-
Building Infectious Disease Research Programs to Promote Security and Enhance Collaborations with Countries of the Former Soviet Union.Front Public Health. 2015 Nov 26;3:271. doi: 10.3389/fpubh.2015.00271. eCollection 2015. Front Public Health. 2015. PMID: 26636067 Free PMC article. Review.
Cited by
-
Challenges and Opportunities in Pathogen Agnostic Sequencing for Public Health Surveillance: Lessons Learned From the Global Emerging Infections Surveillance Program.Health Secur. 2024 Jan-Feb;22(1):16-24. doi: 10.1089/hs.2023.0068. Epub 2023 Dec 6. Health Secur. 2024. PMID: 38054950 Free PMC article. No abstract available.
-
Next-Generation Sequencing and Bioinformatics Consortium Approach to Genomic Surveillance.Emerg Infect Dis. 2024 Oct;30(14):13-18. doi: 10.3201/eid3014.240306. Emerg Infect Dis. 2024. PMID: 39530777 Free PMC article.
References
-
- Centers for Disease Control and Prevention (2019). SARS-CoV-2 Sequencing for Public Health Emergency Response, Epidemiology, and Surveillance 2020. Available online at: https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/spheres.html (accessed August 6, 2020).
-
- Chakhunashvili G., Wagner A. L., Machablishvili A., Karseladze I., Tarkhan-Mouravi O., Zakhashvili K., et al. (2017). Implementation of a sentinel surveillance system for influenza-like illness (ILI) and severe acute respiratory infection (SARI) in the country of Georgia, 2015-2016. Int. J. Infect. Dis. 65 98–100. 10.1016/j.ijid.2017.09.028 - DOI - PubMed
-
- Chang K. S., Kim G. H., Ha Y. R., Jeong E. K., Kim H. C., Klein T. A., et al. (2018). Monitoring and control of Aedes albopictus, a vector of Zika Virus, near residences of imported Zika Virus patients during 2016 in South Korea. Am. J. Trop. Med. Hyg. 98 166–172. 10.4269/ajtmh.17-0587 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

