A Survey of Regulatory Interactions Among RNA Binding Proteins and MicroRNAs in Cancer
- PMID: 33101370
- PMCID: PMC7506142
- DOI: 10.3389/fgene.2020.515094
A Survey of Regulatory Interactions Among RNA Binding Proteins and MicroRNAs in Cancer
Abstract
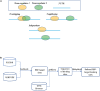
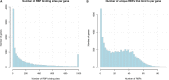
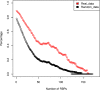
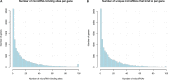
Recent advances in genomics and proteomics generated a large amount of trans regulatory data such as those mediated by RNA binding proteins (RBPs) and microRNAs. Since many trans regulators target 3' UTR of mRNA transcripts, it is likely that there would be interactions, i.e., competitive or cooperative effect, among these trans factors. We compiled the available RBP and microRNA binding sites, mapped them to the mRNA transcripts, and correlated the binding data with mRNA expression data generated by The Cancer Genome Atlas (TCGA). We separated pairs of RBPs and microRNAs into three scenarios: those that have overlapping target sites on the same mRNA transcript (overlapping), those that have target sites on the same mRNA transcript but non-overlapping (neighboring), and those that do not target the same mRNA transcript (independent). Through a regression analysis on expression profiles, we indeed observed interaction effect between RBPs and microRNAs in the majority of the cancer expression data sets. We further discussed implication of such widespread interactions in the context of cancer and diseases.
Keywords: RNA binding proteins; cancer; gene expression; gene regulation; microRNAs; regression analysis.
Copyright © 2020 Liu, Pan, Kong, Luo and Zhang.
Figures


Similar articles
-
Functional interactions between microRNAs and RNA binding proteins.Microrna. 2012;1(1):70-9. doi: 10.2174/2211536611201010070. Microrna. 2012. PMID: 25048093 Free PMC article. Review.
-
Dissecting the expression relationships between RNA-binding proteins and their cognate targets in eukaryotic post-transcriptional regulatory networks.Sci Rep. 2016 May 10;6:25711. doi: 10.1038/srep25711. Sci Rep. 2016. PMID: 27161996 Free PMC article.
-
SimiRa: A tool to identify coregulation between microRNAs and RNA-binding proteins.RNA Biol. 2015;12(9):998-1009. doi: 10.1080/15476286.2015.1068496. RNA Biol. 2015. PMID: 26383775 Free PMC article.
-
Modeling the combined effect of RNA-binding proteins and microRNAs in post-transcriptional regulation.Nucleic Acids Res. 2016 May 19;44(9):e83. doi: 10.1093/nar/gkw048. Epub 2016 Feb 2. Nucleic Acids Res. 2016. PMID: 26837572 Free PMC article.
-
Competing Interactions of RNA-Binding Proteins, MicroRNAs, and Their Targets Control Neuronal Development and Function.Biomolecules. 2015 Oct 23;5(4):2903-18. doi: 10.3390/biom5042903. Biomolecules. 2015. PMID: 26512708 Free PMC article. Review.
Cited by
-
Inferring RNA-binding protein target preferences using adversarial domain adaptation.PLoS Comput Biol. 2022 Feb 24;18(2):e1009863. doi: 10.1371/journal.pcbi.1009863. eCollection 2022 Feb. PLoS Comput Biol. 2022. PMID: 35202389 Free PMC article.
References
LinkOut - more resources
Full Text Sources

