SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling
- PMID: 33097660
- PMCID: PMC7668094
- DOI: 10.1073/pnas.2016650117
SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling
Abstract
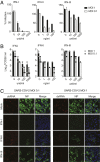
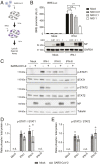
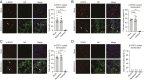
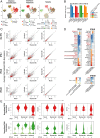
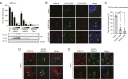
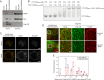
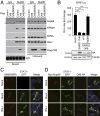
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic that is a serious global health problem. Evasion of IFN-mediated antiviral signaling is a common defense strategy that pathogenic viruses use to replicate and propagate in their host. In this study, we show that SARS-CoV-2 is able to efficiently block STAT1 and STAT2 nuclear translocation in order to impair transcriptional induction of IFN-stimulated genes (ISGs). Our results demonstrate that the viral accessory protein Orf6 exerts this anti-IFN activity. We found that SARS-CoV-2 Orf6 localizes at the nuclear pore complex (NPC) and directly interacts with Nup98-Rae1 via its C-terminal domain to impair docking of cargo-receptor (karyopherin/importin) complex and disrupt nuclear import. In addition, we show that a methionine-to-arginine substitution at residue 58 impairs Orf6 binding to the Nup98-Rae1 complex and abolishes its IFN antagonistic function. All together our data unravel a mechanism of viral antagonism in which a virus hijacks the Nup98-Rae1 complex to overcome the antiviral action of IFN.
Keywords: Nup98; ORF6; SARS-CoV-2; STATs; interferon signaling antagonism.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98.mBio. 2021 Apr 13;12(2):e00065-21. doi: 10.1128/mBio.00065-21. mBio. 2021. PMID: 33849972 Free PMC article.
-
SARS-CoV-2 Orf6 is positioned in the nuclear pore complex by Rae1 to inhibit nucleocytoplasmic transport.Mol Biol Cell. 2024 May 1;35(5):ar62. doi: 10.1091/mbc.E23-10-0386. Epub 2024 Mar 20. Mol Biol Cell. 2024. PMID: 38507240 Free PMC article.
-
Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex.Biochem Biophys Res Commun. 2021 Jan 15;536:59-66. doi: 10.1016/j.bbrc.2020.11.115. Epub 2020 Dec 13. Biochem Biophys Res Commun. 2021. PMID: 33360543 Free PMC article.
-
Roles and functions of SARS-CoV-2 proteins in host immune evasion.Front Immunol. 2022 Aug 8;13:940756. doi: 10.3389/fimmu.2022.940756. eCollection 2022. Front Immunol. 2022. PMID: 36003396 Free PMC article. Review.
-
Evasion of interferon responses by Ebola and Marburg viruses.J Interferon Cytokine Res. 2009 Sep;29(9):511-20. doi: 10.1089/jir.2009.0076. J Interferon Cytokine Res. 2009. PMID: 19694547 Free PMC article. Review.
Cited by
-
MDA5 Is a Major Determinant of Developing Symptoms in Critically Ill COVID-19 Patients.Clin Rev Allergy Immunol. 2024 Dec;67(1-3):58-72. doi: 10.1007/s12016-024-09008-z. Epub 2024 Oct 26. Clin Rev Allergy Immunol. 2024. PMID: 39460899 Review.
-
SARS-CoV-2 infects human adult donor eyes and hESC-derived ocular epithelium.Cell Stem Cell. 2021 Jul 1;28(7):1205-1220.e7. doi: 10.1016/j.stem.2021.04.028. Epub 2021 May 17. Cell Stem Cell. 2021. PMID: 34022129 Free PMC article.
-
Nanomaterials to combat SARS-CoV-2: Strategies to prevent, diagnose and treat COVID-19.Front Bioeng Biotechnol. 2022 Nov 25;10:1052436. doi: 10.3389/fbioe.2022.1052436. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36507266 Free PMC article. Review.
-
SARS-CoV-2 viral protein ORF3A injures renal tubules by interacting with TRIM59 to induce STAT3 activation.Mol Ther. 2023 Mar 1;31(3):774-787. doi: 10.1016/j.ymthe.2022.12.008. Epub 2022 Dec 15. Mol Ther. 2023. PMID: 36523164 Free PMC article.
-
Translational Control of COVID-19 and Its Therapeutic Implication.Front Immunol. 2022 Mar 29;13:857490. doi: 10.3389/fimmu.2022.857490. eCollection 2022. Front Immunol. 2022. PMID: 35422818 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

