Genome-wide study of pineapple (Ananas comosus L.) bHLH transcription factors indicates that cryptochrome-interacting bHLH2 (AcCIB2) participates in flowering time regulation and abiotic stress response
- PMID: 33092537
- PMCID: PMC7583237
- DOI: 10.1186/s12864-020-07152-2
Genome-wide study of pineapple (Ananas comosus L.) bHLH transcription factors indicates that cryptochrome-interacting bHLH2 (AcCIB2) participates in flowering time regulation and abiotic stress response
Abstract
Background: Transcription factors (TFs) are essential regulators of growth and development in eukaryotes. Basic-helix-loop-helix (bHLHs) is one of the most significant TFs families involved in several critical regulatory functions. Cryptochrome-interacting bHLH (CIB) and cryptochromes form an extensive regulatory network to mediate a plethora of pathways. Although bHLHs regulate critical biological processes in plants, the information about pineapple bHLHs remains unexplored.
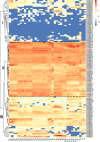
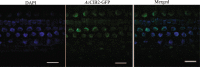
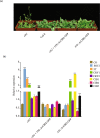
Results: Here, we identified a total of 121 bHLH proteins in the pineapple genome. The identified genes were renamed based on the ascending order of their gene ID and classified into 18 subgroups by phylogenetic analysis. We found that bHLH genes are expressed in different organs and stages of pineapple development. Furthermore, by the ectopic expression of AcCIB2 in Arabidopsis and complementation of Atcib2 mutant, we verified the involvement of AcCIB2 in photomorphogenesis and abiotic stress response.
Conclusions: Our findings revealed that AcCIB2 plays an essential role in flowering time regulation and abiotic stress response. The present study provides additional insights into the current knowledge of bHLH genes and suggests their potential role in various biological processes during pineapple development.
Keywords: Abiotic stress; CIB2; Flowering time; Pineapple; bHLH.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus).BMC Genomics. 2017 Jul 1;18(1):503. doi: 10.1186/s12864-017-3896-y. BMC Genomics. 2017. PMID: 28668094 Free PMC article.
-
Current Understanding of bHLH Transcription Factors in Plant Abiotic Stress Tolerance.Int J Mol Sci. 2021 May 6;22(9):4921. doi: 10.3390/ijms22094921. Int J Mol Sci. 2021. PMID: 34066424 Free PMC article. Review.
-
Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress.BMC Genomics. 2018 Jun 25;19(1):490. doi: 10.1186/s12864-018-4880-x. BMC Genomics. 2018. PMID: 29940851 Free PMC article.
-
Genome-Wide Identification and Expression Profiling of CBL-CIPK Gene Family in Pineapple (Ananas comosus) and the Role of AcCBL1 in Abiotic and Biotic Stress Response.Biomolecules. 2019 Jul 20;9(7):293. doi: 10.3390/biom9070293. Biomolecules. 2019. PMID: 31330847 Free PMC article.
-
Basic Helix-Loop-Helix Transcription Factors: Regulators for Plant Growth Development and Abiotic Stress Responses.Int J Mol Sci. 2023 Jan 11;24(2):1419. doi: 10.3390/ijms24021419. Int J Mol Sci. 2023. PMID: 36674933 Free PMC article. Review.
Cited by
-
Genome-wide characterization and expression analysis of bHLH gene family in physic nut (Jatropha curcas L.).PeerJ. 2022 Aug 9;10:e13786. doi: 10.7717/peerj.13786. eCollection 2022. PeerJ. 2022. PMID: 35966923 Free PMC article.
-
Integrative Analyses of Transcriptomes and Metabolomes Reveal Associated Genes and Metabolites with Flowering Regulation in Common Vetch (Vicia sativa L.).Int J Mol Sci. 2022 Jun 19;23(12):6818. doi: 10.3390/ijms23126818. Int J Mol Sci. 2022. PMID: 35743262 Free PMC article.
-
Integrated Analysis of Basic Helix Loop Helix Transcription Factor Family and Targeted Terpenoids Reveals Candidate AarbHLH Genes Involved in Terpenoid Biosynthesis in Artemisia argyi.Front Plant Sci. 2022 Jan 17;12:811166. doi: 10.3389/fpls.2021.811166. eCollection 2021. Front Plant Sci. 2022. PMID: 35111184 Free PMC article.
-
Genome-Wide Characterization of bHLH Family Genes and Expression Analysis in Response to Osmotic Stress in Betula platyphylla.Plants (Basel). 2023 Oct 25;12(21):3687. doi: 10.3390/plants12213687. Plants (Basel). 2023. PMID: 37960044 Free PMC article.
-
AtCRY2 Negatively Regulates the Functions of AtANN2 and AtANN3 in Drought Tolerance by Affecting Their Subcellular Localization and Transmembrane Ca2+ Flow.Front Plant Sci. 2021 Nov 23;12:754567. doi: 10.3389/fpls.2021.754567. eCollection 2021. Front Plant Sci. 2021. PMID: 34887887 Free PMC article.
References
-
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153(3):1398–1412. doi: 10.1104/pp.110.153593. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

