Down-regulation of Beclin1 promotes direct cardiac reprogramming
- PMID: 33087505
- PMCID: PMC8188650
- DOI: 10.1126/scitranslmed.aay7856
Down-regulation of Beclin1 promotes direct cardiac reprogramming
Abstract
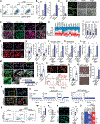
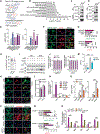
Direct reprogramming of fibroblasts to alternative cell fates by forced expression of transcription factors offers a platform to explore fundamental molecular events governing cell fate identity. The discovery and study of induced cardiomyocytes (iCMs) not only provides alternative therapeutic strategies for heart disease but also sheds lights on basic biology underlying CM fate determination. The iCM field has primarily focused on early transcriptome and epigenome repatterning, whereas little is known about how reprogramming iCMs remodel, erase, and exit the initial fibroblast lineage to acquire final cell identity. Here, we show that autophagy-related 5 (Atg5)-dependent autophagy, an evolutionarily conserved self-digestion process, was induced and required for iCM reprogramming. Unexpectedly, the autophagic factor Beclin1 (Becn1) was found to suppress iCM induction in an autophagy-independent manner. Depletion of Becn1 resulted in improved iCM induction from both murine and human fibroblasts. In a mouse genetic model, Becn1 haploinsufficiency further enhanced reprogramming factor-mediated heart function recovery and scar size reduction after myocardial infarction. Mechanistically, loss of Becn1 up-regulated Lef1 and down-regulated Wnt inhibitors, leading to activation of the canonical Wnt/β-catenin signaling pathway. In addition, Becn1 physically interacts with other classical class III phosphatidylinositol 3-kinase (PI3K III) complex components, the knockdown of which phenocopied Becn1 depletion in cardiac reprogramming. Collectively, our study revealed an inductive role of Atg5-dependent autophagy as well as a previously unrecognized autophagy-independent inhibitory function of Becn1 in iCM reprogramming.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures






Similar articles
-
Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte.Nature. 2017 Nov 2;551(7678):100-104. doi: 10.1038/nature24454. Epub 2017 Oct 25. Nature. 2017. PMID: 29072293 Free PMC article.
-
Single cell qPCR reveals that additional HAND2 and microRNA-1 facilitate the early reprogramming progress of seven-factor-induced human myocytes.PLoS One. 2017 Aug 10;12(8):e0183000. doi: 10.1371/journal.pone.0183000. eCollection 2017. PLoS One. 2017. PMID: 28796841 Free PMC article.
-
KLF2 (kruppel-like factor 2 [lung]) regulates osteoclastogenesis by modulating autophagy.Autophagy. 2019 Dec;15(12):2063-2075. doi: 10.1080/15548627.2019.1596491. Epub 2019 Apr 16. Autophagy. 2019. PMID: 30894058 Free PMC article.
-
The heart of cardiac reprogramming: The cardiac fibroblasts.J Mol Cell Cardiol. 2022 Nov;172:90-99. doi: 10.1016/j.yjmcc.2022.08.004. Epub 2022 Aug 22. J Mol Cell Cardiol. 2022. PMID: 36007393 Review.
-
Reprogramming of Non-myocytes into Cardiomyocyte-like Cells: Challenges and Opportunities.Curr Cardiol Rep. 2020 Jun 19;22(8):54. doi: 10.1007/s11886-020-01322-0. Curr Cardiol Rep. 2020. PMID: 32562156 Review.
Cited by
-
Identification of molecular subtypes of ischaemic stroke based on immune-related genes and weighted co-expression network analysis.IET Syst Biol. 2023 Apr;17(2):58-69. doi: 10.1049/syb2.12059. Epub 2023 Feb 18. IET Syst Biol. 2023. PMID: 36802116 Free PMC article.
-
p63 silencing induces epigenetic modulation to enhance human cardiac fibroblast to cardiomyocyte-like differentiation.Sci Rep. 2022 Jul 6;12(1):11416. doi: 10.1038/s41598-022-15559-y. Sci Rep. 2022. PMID: 35794145 Free PMC article.
-
Regeneration of the heart: from molecular mechanisms to clinical therapeutics.Mil Med Res. 2023 Apr 26;10(1):18. doi: 10.1186/s40779-023-00452-0. Mil Med Res. 2023. PMID: 37098604 Free PMC article. Review.
-
Cardiac regeneration by direct reprogramming in this decade and beyond.Inflamm Regen. 2021 Jul 1;41(1):20. doi: 10.1186/s41232-021-00168-5. Inflamm Regen. 2021. PMID: 34193320 Free PMC article. Review.
-
Direct reprogramming as a route to cardiac repair.Semin Cell Dev Biol. 2022 Feb;122:3-13. doi: 10.1016/j.semcdb.2021.05.019. Epub 2021 Jul 8. Semin Cell Dev Biol. 2022. PMID: 34246567 Free PMC article. Review.
References
-
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S, Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). - PubMed
-
- Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). - PubMed
-
- Song G, Pacher M, Balakrishnan A, Yuan Q, Tsay H-C, Yang D, Reetz J, Brandes S, Dai Z, Pützer BM, Araúzo-Bravo MJ, Steinemann D, Luedde T, Schwabe RF, Manns MP, Schöler HR, Schambach A, Cantz T, Ott M, Sharma AD, Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell 18, 797–808 (2016). - PubMed
-
- Han DW, Greber B, Wu G, Tapia N, Araúzo-Bravo MJ, Ko K, Bernemann C, Stehling M, Schöler HR, Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol 13, 66–71 (2011). - PubMed
-
- Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L, Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

