This is a preprint.
A betacoronavirus multiplex microsphere immunoassay detects early SARS-CoV-2 seroconversion and controls for pre-existing seasonal human coronavirus antibody cross-reactivity
- PMID: 33083807
- PMCID: PMC7574255
- DOI: 10.1101/2020.10.14.20207050
A betacoronavirus multiplex microsphere immunoassay detects early SARS-CoV-2 seroconversion and controls for pre-existing seasonal human coronavirus antibody cross-reactivity
Abstract
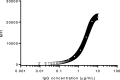
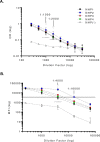
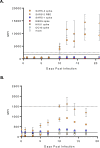
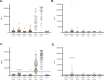
With growing concern of persistent or multiple waves of SARS-CoV-2 in the United States, sensitive and specific SARS-CoV-2 antibody assays remain critical for community and hospital-based SARS-CoV-2 surveillance. Here, we describe the development and application of a multiplex microsphere-based immunoassay (MMIA) for COVD-19 antibody studies, utilizing serum samples from non-human primate SARS-CoV-2 infection models, an archived human sera bank and subjects enrolled at five U.S. military hospitals. The MMIA incorporates prefusion stabilized spike glycoprotein trimers of SARS-CoV-2, SARS-CoV-1, MERS-CoV, and the seasonal human coronaviruses HCoV-HKU1 and HCoV-OC43, into a multiplexing system that enables simultaneous measurement of off-target pre-existing cross-reactive antibodies. We report the sensitivity and specificity performances for this assay strategy at 98% sensitivity and 100% specificity for subject samples collected as early as 10 days after the onset of symptoms. In archival sera collected prior to 2019 and serum samples from subjects PCR negative for SARS-CoV-2, we detected seroprevalence of 72% and 98% for HCoV-HKU1 and HCoV-0C43, respectively. Requiring only 1.25 μL of sera, this approach permitted the simultaneous identification of SARS-CoV-2 seroconversion and polyclonal SARS-CoV-2 IgG antibody responses to SARS-CoV-1 and MERS-CoV, further demonstrating the presence of conserved epitopes in the spike glycoprotein of zoonotic betacoronaviruses. Application of this serology assay in observational studies with serum samples collected from subjects before and after SARS-CoV-2 infection will permit an investigation of the influences of HCoV-induced antibodies on COVID-19 clinical outcomes.
Conflict of interest statement
CONFLICT OF INTEREST None of the authors have any conflicts of interest of relevance to disclose.
Figures



Similar articles
-
A betacoronavirus multiplex microsphere immunoassay detects early SARS-CoV-2 seroconversion and antibody cross reactions.Res Sq [Preprint]. 2020 Nov 24:rs.3.rs-105768. doi: 10.21203/rs.3.rs-105768/v1. Res Sq. 2020. PMID: 33269345 Free PMC article. Preprint.
-
Antigen-based multiplex strategies to discriminate SARS-CoV-2 natural and vaccine induced immunity from seasonal human coronavirus humoral responses.medRxiv [Preprint]. 2021 Feb 12:2021.02.10.21251518. doi: 10.1101/2021.02.10.21251518. medRxiv. 2021. PMID: 33594376 Free PMC article. Preprint.
-
Prospective Assessment of SARS-CoV-2 Seroconversion (PASS) study: an observational cohort study of SARS-CoV-2 infection and vaccination in healthcare workers.BMC Infect Dis. 2021 Jun 9;21(1):544. doi: 10.1186/s12879-021-06233-1. BMC Infect Dis. 2021. PMID: 34107889 Free PMC article.
-
Roles of Sialyl Glycans in HCoV-OC43, HCoV-HKU1, MERS-CoV and SARS-CoV-2 Infections.Methods Mol Biol. 2022;2556:243-271. doi: 10.1007/978-1-0716-2635-1_17. Methods Mol Biol. 2022. PMID: 36175638 Review.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
References
-
- Long Q. X. et al., Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 26, 1200–1204 (2020). - PubMed
-
- Seow J. et al., Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv 10.1101/2020.07.09.20148429, 2020.2007.2009.20148429 (2020). - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
