Mechanism of Type IA Topoisomerases
- PMID: 33080770
- PMCID: PMC7587558
- DOI: 10.3390/molecules25204769
Mechanism of Type IA Topoisomerases
Abstract
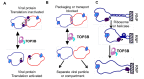
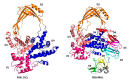
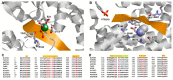
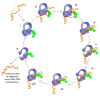
Topoisomerases in the type IA subfamily can catalyze change in topology for both DNA and RNA substrates. A type IA topoisomerase may have been present in a last universal common ancestor (LUCA) with an RNA genome. Type IA topoisomerases have since evolved to catalyze the resolution of topological barriers encountered by genomes that require the passing of nucleic acid strand(s) through a break on a single DNA or RNA strand. Here, based on available structural and biochemical data, we discuss how a type IA topoisomerase may recognize and bind single-stranded DNA or RNA to initiate its required catalytic function. Active site residues assist in the nucleophilic attack of a phosphodiester bond between two nucleotides to form a covalent intermediate with a 5'-phosphotyrosine linkage to the cleaved nucleic acid. A divalent ion interaction helps to position the 3'-hydroxyl group at the precise location required for the cleaved phosphodiester bond to be rejoined following the passage of another nucleic acid strand through the break. In addition to type IA topoisomerase structures observed by X-ray crystallography, we now have evidence from biophysical studies for the dynamic conformations that are required for type IA topoisomerases to catalyze the change in the topology of the nucleic acid substrates.
Keywords: DNA supercoiling; RNA topology; decatenation; topoisomerase; type IA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule.Nature. 2001 Jun 28;411(6841):1077-81. doi: 10.1038/35082615. Nature. 2001. PMID: 11429611
-
The type IA topoisomerase catalytic cycle: A normal mode analysis and molecular dynamics simulation.Proteins. 2008 Jun;71(4):1984-94. doi: 10.1002/prot.21876. Proteins. 2008. PMID: 18186484
-
The structure of Escherichia coli DNA topoisomerase III.Structure. 1999 Nov 15;7(11):1373-83. doi: 10.1016/s0969-2126(00)80027-1. Structure. 1999. PMID: 10574789
-
What's on the Other Side of the Gate: A Structural Perspective on DNA Gate Opening of Type IA and IIA DNA Topoisomerases.Int J Mol Sci. 2023 Feb 16;24(4):3986. doi: 10.3390/ijms24043986. Int J Mol Sci. 2023. PMID: 36835394 Free PMC article. Review.
-
Helicase-appended topoisomerases: new insight into the mechanism of directional strand transfer.J Biol Chem. 2009 Nov 6;284(45):30737-41. doi: 10.1074/jbc.R109.051268. Epub 2009 Sep 2. J Biol Chem. 2009. PMID: 19726668 Free PMC article. Review.
Cited by
-
The interaction between transport-segment DNA and topoisomerase IA-crystal structure of MtbTOP1 in complex with both G- and T-segments.Nucleic Acids Res. 2023 Jan 11;51(1):349-364. doi: 10.1093/nar/gkac1205. Nucleic Acids Res. 2023. PMID: 36583363 Free PMC article.
-
Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life.Cells. 2024 Mar 21;13(6):553. doi: 10.3390/cells13060553. Cells. 2024. PMID: 38534397 Free PMC article. Review.
-
SUMO: A Swiss Army Knife for Eukaryotic Topoisomerases.Front Mol Biosci. 2022 Apr 6;9:871161. doi: 10.3389/fmolb.2022.871161. eCollection 2022. Front Mol Biosci. 2022. PMID: 35463961 Free PMC article. Review.
-
Localization of Mycobacterium tuberculosis topoisomerase I C-terminal sequence motif required for inhibition by endogenous toxin MazF4.Front Microbiol. 2022 Dec 5;13:1032320. doi: 10.3389/fmicb.2022.1032320. eCollection 2022. Front Microbiol. 2022. PMID: 36545199 Free PMC article.
-
Enzymatic Processing of DNA-Protein Crosslinks.Genes (Basel). 2024 Jan 10;15(1):85. doi: 10.3390/genes15010085. Genes (Basel). 2024. PMID: 38254974 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

