Soybean DICER-LIKE2 Regulates Seed Coat Color via Production of Primary 22-Nucleotide Small Interfering RNAs from Long Inverted Repeats
- PMID: 33077493
- PMCID: PMC7721327
- DOI: 10.1105/tpc.20.00562
Soybean DICER-LIKE2 Regulates Seed Coat Color via Production of Primary 22-Nucleotide Small Interfering RNAs from Long Inverted Repeats
Abstract
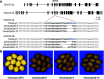
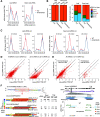
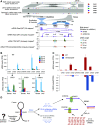
In plants, 22-nucleotide small RNAs trigger the production of secondary small interfering RNAs (siRNAs) and enhance silencing. DICER-LIKE2 (DCL2)-dependent 22-nucleotide siRNAs are rare in Arabidopsis (Arabidopsis thaliana) and are thought to function mainly during viral infection; by contrast, these siRNAs are abundant in many crops such as soybean (Glycine max) and maize (Zea mays). Here, we studied soybean 22-nucleotide siRNAs by applying CRISPR-Cas9 to simultaneously knock out the two copies of soybean DCL2, GmDCL2a and GmDCL2b, in the Tianlong1 cultivar. Small RNA sequencing revealed that most 22-nucleotide siRNAs are derived from long inverted repeats (LIRs) and disappeared in the Gmdcl2a/2b double mutant. De novo assembly of a Tianlong1 reference genome and transcriptome profiling identified an intronic LIR formed by the chalcone synthase (CHS) genes CHS1 and CHS3 This LIR is the source of primary 22-nucleotide siRNAs that target other CHS genes and trigger the production of secondary 21-nucleotide siRNAs. Disruption of this process in Gmdcl2a/2b mutants substantially increased CHS mRNA levels in the seed coat, thus changing the coat color from yellow to brown. Our results demonstrated that endogenous LIR-derived transcripts in soybean are predominantly processed by GmDCL2 into 22-nucleotide siRNAs and uncovered a role for DCL2 in regulating natural traits.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures


Comment in
-
Slice and Dice: DCL2 Mediates the Production of 22-Nucleotide siRNAs that Influence Trait Variation in Soybean.Plant Cell. 2020 Dec;32(12):3646-3647. doi: 10.1105/tpc.20.00884. Epub 2020 Oct 22. Plant Cell. 2020. PMID: 33093146 Free PMC article. No abstract available.
Similar articles
-
The transition from primary siRNAs to amplified secondary siRNAs that regulate chalcone synthase during development of Glycine max seed coats.PLoS One. 2013 Oct 21;8(10):e76954. doi: 10.1371/journal.pone.0076954. eCollection 2013. PLoS One. 2013. PMID: 24204712 Free PMC article.
-
Endogenous RNA interference of chalcone synthase genes in soybean: formation of double-stranded RNA of GmIRCHS transcripts and structure of the 5' and 3' ends of short interfering RNAs.J Plant Physiol. 2011 Jul 15;168(11):1264-70. doi: 10.1016/j.jplph.2011.01.003. Epub 2011 Feb 3. J Plant Physiol. 2011. PMID: 21295373
-
Molecular mechanism of seed coat discoloration induced by low temperature in yellow soybean.Plant Cell Physiol. 2009 Jun;50(6):1090-8. doi: 10.1093/pcp/pcp061. Epub 2009 Apr 23. Plant Cell Physiol. 2009. PMID: 19395413
-
Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in glycine max seed coats.Plant Cell. 2009 Oct;21(10):3063-77. doi: 10.1105/tpc.109.069856. Epub 2009 Oct 9. Plant Cell. 2009. PMID: 19820189 Free PMC article.
-
Biogenesis of trans-acting siRNAs, endogenous secondary siRNAs in plants.Genes Genet Syst. 2013;88(2):77-84. doi: 10.1266/ggs.88.77. Genes Genet Syst. 2013. PMID: 23832299 Review.
Cited by
-
Endogenous Caulimovirids: Fossils, Zombies, and Living in Plant Genomes.Biomolecules. 2023 Jul 3;13(7):1069. doi: 10.3390/biom13071069. Biomolecules. 2023. PMID: 37509105 Free PMC article. Review.
-
Metabolomic and transcriptomic analyses reveal the effects of self- and hetero-grafting on anthocyanin biosynthesis in grapevine.Hortic Res. 2022 May 17;9:uhac103. doi: 10.1093/hr/uhac103. eCollection 2022. Hortic Res. 2022. PMID: 35795384 Free PMC article.
-
Induced Mutation in GmCOP1b Enhances the Performance of Soybean under Dense Planting Conditions.Int J Mol Sci. 2022 May 12;23(10):5394. doi: 10.3390/ijms23105394. Int J Mol Sci. 2022. PMID: 35628205 Free PMC article.
-
Identification of genetic loci conferring seed coat color based on a high-density map in soybean.Front Plant Sci. 2022 Aug 1;13:968618. doi: 10.3389/fpls.2022.968618. eCollection 2022. Front Plant Sci. 2022. PMID: 35979081 Free PMC article.
-
Interspecific hybridization in tomato influences endogenous viral sRNAs and alters gene expression.Genome Biol. 2022 May 21;23(1):120. doi: 10.1186/s13059-022-02685-z. Genome Biol. 2022. PMID: 35597968 Free PMC article.
References
-
- Allen E., Xie Z., Gustafson A.M., Carrington J.C.(2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. - PubMed
-
- Allen E., Xie Z., Gustafson A.M., Sung G.-H., Spatafora J.W., Carrington J.C.(2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290. - PubMed
-
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P.(2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

