Expression of mitochondrial protein genes encoded by nuclear and mitochondrial genomes correlate with energy metabolism in dairy cattle
- PMID: 33076826
- PMCID: PMC7574280
- DOI: 10.1186/s12864-020-07018-7
Expression of mitochondrial protein genes encoded by nuclear and mitochondrial genomes correlate with energy metabolism in dairy cattle
Erratum in
-
Correction to: Expression of mitochondrial protein genes encoded by nuclear and mitochondrial genomes correlate with energy metabolism in dairy cattle.BMC Genomics. 2022 Apr 20;23(1):315. doi: 10.1186/s12864-022-08404-z. BMC Genomics. 2022. PMID: 35443605 Free PMC article. No abstract available.
Abstract
Background: Mutations in the mitochondrial genome have been implicated in mitochondrial disease, often characterized by impaired cellular energy metabolism. Cellular energy metabolism in mitochondria involves mitochondrial proteins (MP) from both the nuclear (NuMP) and mitochondrial (MtMP) genomes. The expression of MP genes in tissues may be tissue specific to meet varying specific energy demands across the tissues. Currently, the characteristics of MP gene expression in tissues of dairy cattle are not well understood. In this study, we profile the expression of MP genes in 29 adult and six foetal tissues in dairy cattle using RNA sequencing and gene expression analyses: particularly differential gene expression and co-expression network analyses.
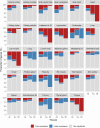
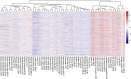
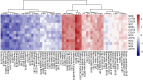
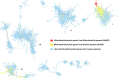
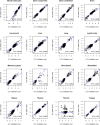
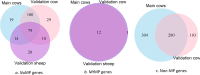
Results: MP genes were differentially expressed (DE; over-expressed or under-expressed) across tissues in cattle. All 29 tissues showed DE NuMP genes in varying proportions of over-expression and under-expression. On the other hand, DE of MtMP genes was observed in < 50% of tissues and notably MtMP genes within a tissue was either all over-expressed or all under-expressed. A high proportion of NuMP (up to 60%) and MtMP (up to 100%) genes were over-expressed in tissues with expected high metabolic demand; heart, skeletal muscles and tongue, and under-expressed (up to 45% of NuMP, 77% of MtMP genes) in tissues with expected low metabolic rates; leukocytes, thymus, and lymph nodes. These tissues also invariably had the expression of all MtMP genes in the direction of dominant NuMP genes expression. The NuMP and MtMP genes were highly co-expressed across tissues and co-expression of genes in a cluster were non-random and functionally enriched for energy generation pathway. The differential gene expression and co-expression patterns were validated in independent cow and sheep datasets.
Conclusions: The results of this study support the concept that there are biological interaction of MP genes from the mitochondrial and nuclear genomes given their over-expression in tissues with high energy demand and co-expression in tissues. This highlights the importance of considering MP genes from both genomes in future studies related to mitochondrial functions and traits related to energy metabolism.
Keywords: Cattle; Differential gene expression; Energy metabolism; Gene co-expression; Mitochondria.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Mitochondrial protein gene expression and the oxidative phosphorylation pathway associated with feed efficiency and energy balance in dairy cattle.J Dairy Sci. 2021 Jan;104(1):575-587. doi: 10.3168/jds.2020-18503. Epub 2020 Nov 6. J Dairy Sci. 2021. PMID: 33162069
-
Differential gene expression in dairy cows under negative energy balance and ketosis: A systematic review and meta-analysis.J Dairy Sci. 2021 Jan;104(1):602-615. doi: 10.3168/jds.2020-18883. Epub 2020 Nov 12. J Dairy Sci. 2021. PMID: 33189279
-
High-density genome-wide association study for residual feed intake in Holstein dairy cattle.J Dairy Sci. 2019 Dec;102(12):11067-11080. doi: 10.3168/jds.2019-16645. Epub 2019 Sep 25. J Dairy Sci. 2019. PMID: 31563317
-
Graduate Student Literature Review: Mitochondrial adaptations across lactation and their molecular regulation in dairy cattle.J Dairy Sci. 2021 Sep;104(9):10415-10425. doi: 10.3168/jds.2021-20138. Epub 2021 Jul 1. J Dairy Sci. 2021. PMID: 34218917 Review.
-
Is mitochondrial gene expression coordinated or stochastic?Biochem Soc Trans. 2018 Oct 19;46(5):1239-1246. doi: 10.1042/BST20180174. Epub 2018 Oct 8. Biochem Soc Trans. 2018. PMID: 30301847 Review.
Cited by
-
Putative Causal Variants Are Enriched in Annotated Functional Regions From Six Bovine Tissues.Front Genet. 2021 Jun 23;12:664379. doi: 10.3389/fgene.2021.664379. eCollection 2021. Front Genet. 2021. PMID: 34249087 Free PMC article.
-
Feather Gene Expression Elucidates the Developmental Basis of Plumage Iridescence in African Starlings.J Hered. 2021 Aug 25;112(5):417-429. doi: 10.1093/jhered/esab014. J Hered. 2021. PMID: 33885791 Free PMC article.
-
AgAnimalGenomes: browsers for viewing and manually annotating farm animal genomes.Mamm Genome. 2023 Sep;34(3):418-436. doi: 10.1007/s00335-023-10008-1. Epub 2023 Jul 17. Mamm Genome. 2023. PMID: 37460664 Free PMC article.
-
Nuclear Genome-Encoded Mitochondrial OXPHOS Complex I Genes in Female Buffalo Show Tissue-Specific Differences.Mol Biotechnol. 2024 Jun 15. doi: 10.1007/s12033-024-01206-6. Online ahead of print. Mol Biotechnol. 2024. PMID: 38878239
-
Correction to: Expression of mitochondrial protein genes encoded by nuclear and mitochondrial genomes correlate with energy metabolism in dairy cattle.BMC Genomics. 2022 Apr 20;23(1):315. doi: 10.1186/s12864-022-08404-z. BMC Genomics. 2022. PMID: 35443605 Free PMC article. No abstract available.
References
-
- Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, et al. Mitochondrial diseases. Nat Rev Dis Prim. 2016;2:16080. - PubMed
-
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. - PubMed
-
- Elia M, Livesey G. Energy expenditure and fuel selection in biological systems: the theory and practice of calculations based on indirect calorimetry and tracer methods. World Rev Nutr Diet. 1992;70:68–131. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

