KRAS K104 modification affects the KRASG12D-GEF interaction and mediates cell growth and motility
- PMID: 33060649
- PMCID: PMC7567070
- DOI: 10.1038/s41598-020-74463-5
KRAS K104 modification affects the KRASG12D-GEF interaction and mediates cell growth and motility
Abstract
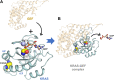
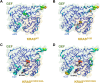
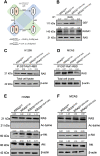
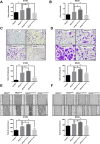
Mutant RAS genes play an important role in regulating tumors through lysine residue 104 to impair GEF-induced nucleotide exchange, but the regulatory role of KRAS K104 modification on the KRASG12D mutant remains unclear. Therefore, we simulated the acetylation site on the KRASG12D three-dimensional protein structure, including KRASG12D, KRASG12D/K104A and KRASG12D/K104Q, and determined their trajectories and binding free energy with GEF. KRASG12D/K104Q induced structural changes in the α2- and α3-helices, promoted KRAS instability and hampered GEF binding (ΔΔG = 6.14 kJ/mol). We found decreased binding to the Raf1 RBD by KRASG12D/K104Q and reduced cell growth, invasion and migration. Based on whole-genome cDNA microarray analysis, KRASG12D/K104Q decreased expression of NPIPA2, DUSP1 and IL6 in lung and ovarian cancer cells. This study reports computational and experimental analyses of Lys104 of KRASG12D and GEF, and the findings provide a target for exploration for future treatment.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.Cancer Res. 2023 Oct 2;83(19):3176-3183. doi: 10.1158/0008-5472.CAN-22-3210. Cancer Res. 2023. PMID: 37556505 Free PMC article.
-
A KRAS GTPase K104Q Mutant Retains Downstream Signaling by Offsetting Defects in Regulation.J Biol Chem. 2017 Mar 17;292(11):4446-4456. doi: 10.1074/jbc.M116.762435. Epub 2017 Jan 30. J Biol Chem. 2017. PMID: 28154176 Free PMC article.
-
Synergistic signaling of KRAS and thyroid hormone receptor β mutants promotes undifferentiated thyroid cancer through MYC up-regulation.Neoplasia. 2014 Sep;16(9):757-69. doi: 10.1016/j.neo.2014.08.003. Neoplasia. 2014. PMID: 25246276 Free PMC article.
-
ETS1 regulates Twist1 transcription in a KrasG12D/Lkb1-/- metastatic lung tumor model of non-small cell lung cancer.Clin Exp Metastasis. 2018 Mar;35(3):149-165. doi: 10.1007/s10585-018-9912-z. Epub 2018 Jun 16. Clin Exp Metastasis. 2018. PMID: 29909489
-
Microsecond Molecular Dynamics Simulation to Gain Insight Into the Binding of MRTX1133 and Trametinib With KRASG12D Mutant Protein for Drug Repurposing.J Mol Recognit. 2024 Nov;37(6):e3103. doi: 10.1002/jmr.3103. Epub 2024 Sep 25. J Mol Recognit. 2024. PMID: 39318275
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

