Transcriptional regulation of MGE progenitor proliferation by PRDM16 controls cortical GABAergic interneuron production
- PMID: 33060132
- PMCID: PMC7687860
- DOI: 10.1242/dev.187526
Transcriptional regulation of MGE progenitor proliferation by PRDM16 controls cortical GABAergic interneuron production
Abstract
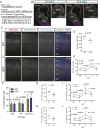
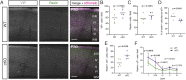
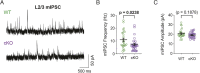
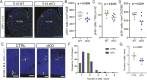
The mammalian cortex is populated by neurons derived from neural progenitors located throughout the embryonic telencephalon. Excitatory neurons are derived from the dorsal telencephalon, whereas inhibitory interneurons are generated in its ventral portion. The transcriptional regulator PRDM16 is expressed by radial glia, neural progenitors present in both regions; however, its mechanisms of action are still not fully understood. It is unclear whether PRDM16 plays a similar role in neurogenesis in both dorsal and ventral progenitor lineages and, if so, whether it regulates common or unique networks of genes. Here, we show that Prdm16 expression in mouse medial ganglionic eminence (MGE) progenitors is required for maintaining their proliferative capacity and for the production of proper numbers of forebrain GABAergic interneurons. PRDM16 binds to cis-regulatory elements and represses the expression of region-specific neuronal differentiation genes, thereby controlling the timing of neuronal maturation. PRDM16 regulates convergent developmental gene expression programs in the cortex and MGE, which utilize both common and region-specific sets of genes to control the proliferative capacity of neural progenitors, ensuring the generation of correct numbers of cortical neurons.
Keywords: CNS development; Cortical interneurons; Medial ganglionic eminence; Neural progenitors; Prdm16.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

Similar articles
-
Non-canonical Wnt Signaling through Ryk Regulates the Generation of Somatostatin- and Parvalbumin-Expressing Cortical Interneurons.Neuron. 2019 Sep 4;103(5):853-864.e4. doi: 10.1016/j.neuron.2019.06.003. Epub 2019 Jun 27. Neuron. 2019. PMID: 31257105 Free PMC article.
-
Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development.Cell Stem Cell. 2013 May 2;12(5):573-86. doi: 10.1016/j.stem.2013.04.005. Cell Stem Cell. 2013. PMID: 23642366 Free PMC article.
-
Prox1 Regulates the Subtype-Specific Development of Caudal Ganglionic Eminence-Derived GABAergic Cortical Interneurons.J Neurosci. 2015 Sep 16;35(37):12869-89. doi: 10.1523/JNEUROSCI.1164-15.2015. J Neurosci. 2015. PMID: 26377473 Free PMC article.
-
Cell cycle regulation and interneuron production.Dev Neurobiol. 2011 Jan 1;71(1):2-9. doi: 10.1002/dneu.20823. Dev Neurobiol. 2011. PMID: 21154905 Free PMC article. Review.
-
Development and Functional Diversification of Cortical Interneurons.Neuron. 2018 Oct 24;100(2):294-313. doi: 10.1016/j.neuron.2018.10.009. Neuron. 2018. PMID: 30359598 Free PMC article. Review.
Cited by
-
Transcriptional profiling of sequentially generated septal neuron fates.Elife. 2021 Dec 1;10:e71545. doi: 10.7554/eLife.71545. Elife. 2021. PMID: 34851821 Free PMC article.
-
PRDM16-DT is a novel lncRNA that regulates astrocyte function in Alzheimer's disease.Acta Neuropathol. 2024 Aug 29;148(1):32. doi: 10.1007/s00401-024-02787-x. Acta Neuropathol. 2024. PMID: 39207536 Free PMC article.
-
Mitochondria in Early Forebrain Development: From Neurulation to Mid-Corticogenesis.Front Cell Dev Biol. 2021 Nov 23;9:780207. doi: 10.3389/fcell.2021.780207. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34888312 Free PMC article. Review.
-
A Single-Cell Transcriptomic Analysis of the Mouse Hippocampus After Voluntary Exercise.Mol Neurobiol. 2024 Aug;61(8):5628-5645. doi: 10.1007/s12035-023-03869-9. Epub 2024 Jan 13. Mol Neurobiol. 2024. PMID: 38217668 Free PMC article.
-
PRDM16 co-operates with LHX2 to shape the human brain.Oxf Open Neurosci. 2024 Jan 24;3:kvae001. doi: 10.1093/oons/kvae001. eCollection 2024. Oxf Open Neurosci. 2024. PMID: 38595939 Free PMC article.
References
-
- Casarosa S., Fode C. and Guillemot F. (1999). Mash1 regulates neurogenesis in the ventral telencephalon. Development 126, 525-534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

