Mechanosensing through Direct Binding of Tensed F-Actin by LIM Domains
- PMID: 33058779
- PMCID: PMC7686152
- DOI: 10.1016/j.devcel.2020.09.022
Mechanosensing through Direct Binding of Tensed F-Actin by LIM Domains
Abstract
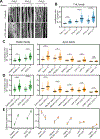
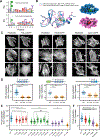
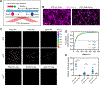
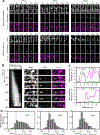
Mechanical signals transmitted through the cytoplasmic actin cytoskeleton must be relayed to the nucleus to control gene expression. LIM domains are protein-protein interaction modules found in cytoskeletal proteins and transcriptional regulators. Here, we identify three LIM protein families (zyxin, paxillin, and FHL) whose members preferentially localize to the actin cytoskeleton in mechanically stimulated cells through their tandem LIM domains. A minimal actin-myosin reconstitution system reveals that representatives of all three families directly bind F-actin only in the presence of mechanical force. Point mutations at a site conserved in each LIM domain of these proteins disrupt tensed F-actin binding in vitro and cytoskeletal localization in cells, demonstrating a common, avidity-based mechanism. Finally, we find that binding to tensed F-actin in the cytoplasm excludes the cancer-associated transcriptional co-activator FHL2 from the nucleus in stiff microenvironments. This establishes direct force-activated F-actin binding as a mechanosensing mechanism by which cytoskeletal tension can govern nuclear localization.
Keywords: FHL2; LIM domain; actin; cytoskeleton; mechanobiology; mechanosensation; mechanotransduction; paxillin; zyxin.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


Similar articles
-
LIM domain proteins in cell mechanobiology.Cytoskeleton (Hoboken). 2021 Jun;78(6):303-311. doi: 10.1002/cm.21677. Epub 2021 Jun 10. Cytoskeleton (Hoboken). 2021. PMID: 34028199 Free PMC article. Review.
-
VASP, zyxin and TES are tension-dependent members of Focal Adherens Junctions independent of the α-catenin-vinculin module.Sci Rep. 2015 Nov 27;5:17225. doi: 10.1038/srep17225. Sci Rep. 2015. PMID: 26611125 Free PMC article.
-
Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25532-25542. doi: 10.1073/pnas.2004656117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989126 Free PMC article.
-
Tandem zyxin LIM sequences do not enhance force sensitive accumulation.Biochem Biophys Res Commun. 2012 Jun 15;422(4):653-7. doi: 10.1016/j.bbrc.2012.05.046. Epub 2012 May 15. Biochem Biophys Res Commun. 2012. PMID: 22609203 Free PMC article.
-
LIM proteins: association with the actin cytoskeleton.Protoplasma. 2002 Feb;219(1-2):1-12. doi: 10.1007/s007090200000. Protoplasma. 2002. PMID: 11926060 Review.
Cited by
-
Mechanosensitive FHL2 tunes endothelial function.bioRxiv [Preprint]. 2024 Jun 17:2024.06.16.599227. doi: 10.1101/2024.06.16.599227. bioRxiv. 2024. PMID: 38948838 Free PMC article. Preprint.
-
Mechanotransduction at the Plasma Membrane-Cytoskeleton Interface.Int J Mol Sci. 2021 Oct 26;22(21):11566. doi: 10.3390/ijms222111566. Int J Mol Sci. 2021. PMID: 34768998 Free PMC article. Review.
-
LIM domain proteins in cell mechanobiology.Cytoskeleton (Hoboken). 2021 Jun;78(6):303-311. doi: 10.1002/cm.21677. Epub 2021 Jun 10. Cytoskeleton (Hoboken). 2021. PMID: 34028199 Free PMC article. Review.
-
Cross-regulations of two connected domains form a mechanical circuit for steady force transmission during clathrin-mediated endocytosis.Cell Rep. 2024 Sep 24;43(9):114725. doi: 10.1016/j.celrep.2024.114725. Epub 2024 Sep 13. Cell Rep. 2024. PMID: 39276354 Free PMC article.
-
Somitic mesoderm morphogenesis is necessary for neural tube closure during Xenopus development.Front Cell Dev Biol. 2023 Jan 9;10:1091629. doi: 10.3389/fcell.2022.1091629. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36699010 Free PMC article.
References
-
- Altman D. (1990). Practical Statistics for Medical Research (Chapman & Hall; ).
-
- Arimura T, Hayashi T, Matsumoto Y, Shibata H, Hiroi S, Nakamura T, Inagaki N, Hinohara K, Takahashi M, Manatsu S-I, et al. (2007). Structural analysis of four and half LIM protein-2 in dilated cardiomyopathy. Biochemical and Biophysical Research Communications 357, 162–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

