Design of novel multiepitope constructs-based peptide vaccine against the structural S, N and M proteins of human COVID-19 using immunoinformatics analysis
- PMID: 33057358
- PMCID: PMC7561160
- DOI: 10.1371/journal.pone.0240577
Design of novel multiepitope constructs-based peptide vaccine against the structural S, N and M proteins of human COVID-19 using immunoinformatics analysis
Abstract
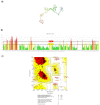
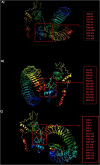
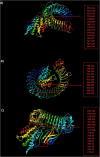
The causative agent of severe acute respiratory syndrome (SARS) reported by the Chinese Center for Disease Control (China CDC) has been identified as a novel Betacoronavirus (SARS-CoV-2). A computational approach was adopted to identify multiepitope vaccine candidates against SARS-CoV-2 based on S, N and M proteins being able to elicit both humoral and cellular immune responses. In this study, the sequence of the virus was obtained from NCBI database and analyzed with in silico tools such as NetMHCpan, IEDB, BepiPred, NetCTL, Tap transport/proteasomal cleavage, Pa3P, GalexyPepDock, I-TASSER, Ellipro and ClusPro. To identify the most immunodominant regions, after analysis of population coverage and epitope conservancy, we proposed three different constructs based on linear B-cell, CTL and HTL epitopes. The 3D structure of constructs was assessed to find discontinuous B-cell epitopes. Among CTL predicted epitopes, S257-265, S603-611 and S360-368, and among HTL predicted epitopes, N167-181, S313-330 and S1110-1126 had better MHC binding rank. We found one putative CTL epitope, S360-368 related to receptor-binding domain (RBD) region for S protein. The predicted epitopes were non-allergen and showed a high quality of proteasomal cleavage and Tap transport efficiency and 100% conservancy within four different clades of SARS-CoV-2. For CTL and HTL epitopes, the highest population coverage of the world's population was calculated for S27-37 with 86.27% and for S196-231, S303-323, S313-330, S1009-1030 and N328-349 with 90.33%, respectively. We identified overall 10 discontinuous B-cell epitopes for three multiepitope constructs. All three constructs showed strong interactions with TLRs 2, 3 and 4 supporting the hypothesis of SARS-CoV-2 susceptibility to TLRs 2, 3 and 4 like other Coronaviridae families. These data demonstrated that the novel designed multiepitope constructs can contribute to develop SARS-CoV-2 peptide vaccine candidates. The in vivo studies are underway using several vaccination strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2.Infect Dis Poverty. 2020 Sep 16;9(1):132. doi: 10.1186/s40249-020-00752-w. Infect Dis Poverty. 2020. PMID: 32938504 Free PMC article.
-
Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2.Virus Res. 2020 Oct 15;288:198082. doi: 10.1016/j.virusres.2020.198082. Epub 2020 Jul 1. Virus Res. 2020. PMID: 32621841 Free PMC article.
-
Designing a multi-epitope peptide based vaccine against SARS-CoV-2.Sci Rep. 2020 Oct 1;10(1):16219. doi: 10.1038/s41598-020-73371-y. Sci Rep. 2020. PMID: 33004978 Free PMC article.
-
Identification of Novel Candidate Epitopes on SARS-CoV-2 Proteins for South America: A Review of HLA Frequencies by Country.Front Immunol. 2020 Sep 3;11:2008. doi: 10.3389/fimmu.2020.02008. eCollection 2020. Front Immunol. 2020. PMID: 33013857 Free PMC article. Review.
-
The SARS-CoV-2 Spike Glycoprotein as a Drug and Vaccine Target: Structural Insights into Its Complexes with ACE2 and Antibodies.Cells. 2020 Oct 22;9(11):2343. doi: 10.3390/cells9112343. Cells. 2020. PMID: 33105869 Free PMC article. Review.
Cited by
-
Insights into structural vaccinology harnessed for universal coronavirus vaccine development.Clin Exp Vaccine Res. 2024 Jul;13(3):202-217. doi: 10.7774/cevr.2024.13.3.202. Epub 2024 Jul 31. Clin Exp Vaccine Res. 2024. PMID: 39144127 Free PMC article. Review.
-
Recombinant vaccines in 2022: a perspective from the cell factory.Microb Cell Fact. 2022 Oct 5;21(1):203. doi: 10.1186/s12934-022-01929-8. Microb Cell Fact. 2022. PMID: 36199085 Free PMC article. Review.
-
An in silico approach to study the role of epitope order in the multi-epitope-based peptide (MEBP) vaccine design.Sci Rep. 2022 Jul 22;12(1):12584. doi: 10.1038/s41598-022-16445-3. Sci Rep. 2022. PMID: 35869117 Free PMC article.
-
Immunoinformatics-guided design of a multi-epitope vaccine based on the structural proteins of severe acute respiratory syndrome coronavirus 2.RSC Adv. 2021 May 19;11(29):18103-18121. doi: 10.1039/d1ra02885e. eCollection 2021 May 13. RSC Adv. 2021. PMID: 35480208 Free PMC article.
-
Identification of vaccine candidate against Omicron variant of SARS-CoV-2 using immunoinformatic approaches.In Silico Pharmacol. 2022 Jul 26;10(1):12. doi: 10.1007/s40203-022-00128-y. eCollection 2022. In Silico Pharmacol. 2022. PMID: 35898574 Free PMC article.
References
-
- Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020; 38: 1–9 - PubMed
-
- Hamming I, Timens W, Bulthuis M, Lely A, Navis Gv, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2004; 203(2): 631–637. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

