The Interaction of Munc18-1 Helix 11 and 12 with the Central Region of the VAMP2 SNARE Motif Is Essential for SNARE Templating and Synaptic Transmission
- PMID: 33055194
- PMCID: PMC7768276
- DOI: 10.1523/ENEURO.0278-20.2020
The Interaction of Munc18-1 Helix 11 and 12 with the Central Region of the VAMP2 SNARE Motif Is Essential for SNARE Templating and Synaptic Transmission
Abstract
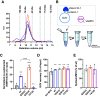
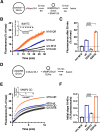
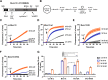
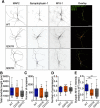
Sec1/Munc18 proteins play a key role in initiating the assembly of N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complexes, the molecular fusion machinery. Employing comparative structure modeling, site specific crosslinking by single amino acid substitutions with the photoactivatable unnatural amino acid p-Benzoyl-phenylalanine (Bpa) and reconstituted vesicle docking/fusion assays, we mapped the binding interface between Munc18-1 and the neuronal v-SNARE VAMP2 with single amino acid resolution. Our results show that helices 11 and 12 of domain 3a in Munc18-1 interact with the VAMP2 SNARE motif covering the region from layers -4 to +5. Residue Q301 in helix 11 plays a pivotal role in VAMP2 binding and template complex formation. A VAMP2 binding deficient mutant, Munc18-1 Q301D, does not stimulate lipid mixing in a reconstituted fusion assay. The neuronal SNARE-organizer Munc13-1, which also binds VAMP2, does not bypass the requirement for the Munc18-1·VAMP2 interaction. Importantly, Munc18-1 Q301D expression in Munc18-1 deficient neurons severely reduces synaptic transmission, demonstrating the physiological significance of the Munc18-1·VAMP2 interaction.
Keywords: Munc18-1; SNARE; VAMP2; crosslinking; membrane fusion; neurotransmission.
Copyright © 2020 André et al.
Figures


Similar articles
-
Energetics, kinetics, and pathways of SNARE assembly in membrane fusion.Crit Rev Biochem Mol Biol. 2022 Aug;57(4):443-460. doi: 10.1080/10409238.2022.2121804. Epub 2022 Sep 24. Crit Rev Biochem Mol Biol. 2022. PMID: 36151854 Free PMC article. Review.
-
An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly.J Biol Chem. 2014 Apr 4;289(14):9639-50. doi: 10.1074/jbc.M113.514273. Epub 2014 Feb 14. J Biol Chem. 2014. PMID: 24532794 Free PMC article.
-
Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association.Elife. 2018 Dec 12;7:e41771. doi: 10.7554/eLife.41771. Elife. 2018. PMID: 30540253 Free PMC article.
-
Tyrosine phosphorylation of Munc18-1 inhibits synaptic transmission by preventing SNARE assembly.EMBO J. 2018 Jan 17;37(2):300-320. doi: 10.15252/embj.201796484. Epub 2017 Nov 17. EMBO J. 2018. PMID: 29150433 Free PMC article.
-
Neuronal SNARE complex assembly guided by Munc18-1 and Munc13-1.FEBS Open Bio. 2022 Nov;12(11):1939-1957. doi: 10.1002/2211-5463.13394. Epub 2022 Mar 22. FEBS Open Bio. 2022. PMID: 35278279 Free PMC article. Review.
Cited by
-
SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.Sci Adv. 2022 Jun 24;8(25):eabo5272. doi: 10.1126/sciadv.abo5272. Epub 2022 Jun 22. Sci Adv. 2022. PMID: 35731863 Free PMC article.
-
SNARE Proteins in Synaptic Vesicle Fusion.Adv Neurobiol. 2023;33:63-118. doi: 10.1007/978-3-031-34229-5_4. Adv Neurobiol. 2023. PMID: 37615864
-
Double NPY motifs at the N-terminus of the yeast t-SNARE Sso2 synergistically bind Sec3 to promote membrane fusion.Elife. 2022 Aug 18;11:e82041. doi: 10.7554/eLife.82041. Elife. 2022. PMID: 35979953 Free PMC article.
-
Energetics, kinetics, and pathways of SNARE assembly in membrane fusion.Crit Rev Biochem Mol Biol. 2022 Aug;57(4):443-460. doi: 10.1080/10409238.2022.2121804. Epub 2022 Sep 24. Crit Rev Biochem Mol Biol. 2022. PMID: 36151854 Free PMC article. Review.
-
Chaperoning SNARE Folding and Assembly.Annu Rev Biochem. 2021 Jun 20;90:581-603. doi: 10.1146/annurev-biochem-081820-103615. Epub 2021 Apr 6. Annu Rev Biochem. 2021. PMID: 33823650 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
