WTAP Function in Sertoli Cells Is Essential for Sustaining the Spermatogonial Stem Cell Niche
- PMID: 33053361
- PMCID: PMC7566211
- DOI: 10.1016/j.stemcr.2020.09.001
WTAP Function in Sertoli Cells Is Essential for Sustaining the Spermatogonial Stem Cell Niche
Abstract
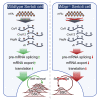
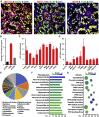
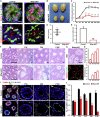
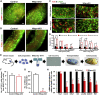
Sertoli cells are the major component of the spermatogonial stem cell (SSC) niche; however, regulatory mechanisms in Sertoli cells that dictate SSC fate decisions remain largely unknown. Here we revealed features of the N6-methyladenosine (m6A) mRNA modification in Sertoli cells and demonstrated the functions of WTAP, the key subunit of the m6A methyltransferase complex in spermatogenesis. m6A-sequencing analysis identified 21,909 m6A sites from 15,365 putative m6A-enriched transcripts within 6,122 genes, including many Sertoli cell-specific genes. Conditional deletion of Wtap in Sertoli cells resulted in sterility and the progressive loss of the SSC population. RNA sequencing and ribosome nascent-chain complex-bound mRNA sequencing analyses suggested that alternative splicing events of transcripts encoding SSC niche factors were sharply altered and translation of these transcripts were severely dysregulated by Wtap deletion. Collectively, this study uncovers a novel regulatory mechanism of the SSC niche and provide insights into molecular interactions between stem cells and their cognate niches in mammals.
Keywords: Sertoli cell; WTAP; alternative splicing; m(6)A; niche; spermatogonial differentiation; spermatogonial stem cell.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling.Oncotarget. 2015 Nov 10;6(35):37012-27. doi: 10.18632/oncotarget.6115. Oncotarget. 2015. PMID: 26473289 Free PMC article.
-
AIP1-mediated actin disassembly is required for postnatal germ cell migration and spermatogonial stem cell niche establishment.Cell Death Dis. 2015 Jul 16;6(7):e1818. doi: 10.1038/cddis.2015.182. Cell Death Dis. 2015. PMID: 26181199 Free PMC article.
-
Temporal-Spatial Establishment of Initial Niche for the Primary Spermatogonial Stem Cell Formation Is Determined by an ARID4B Regulatory Network.Stem Cells. 2017 Jun;35(6):1554-1565. doi: 10.1002/stem.2597. Epub 2017 Mar 16. Stem Cells. 2017. PMID: 28207192 Free PMC article.
-
Molecular regulation of spermatogonial stem cell renewal and differentiation.Reproduction. 2019 Nov;158(5):R169-R187. doi: 10.1530/REP-18-0476. Reproduction. 2019. PMID: 31247585 Review.
-
Regulation of GDNF expression in Sertoli cells.Reproduction. 2019 Mar;157(3):R95-R107. doi: 10.1530/REP-18-0239. Reproduction. 2019. PMID: 30620720 Free PMC article. Review.
Cited by
-
Single-Cell Transcriptomics-Based Study of Transcriptional Regulatory Features in the Non-Obstructive Azoospermia Testis.Front Genet. 2022 May 20;13:875762. doi: 10.3389/fgene.2022.875762. eCollection 2022. Front Genet. 2022. PMID: 35669193 Free PMC article.
-
Ganoderma lucidum Polysaccharide Peptide Alleviates Cyclophosphamide-Induced Male Reproductive Injury by Reducing Oxidative Stress and Apoptosis.Biomedicines. 2024 Jul 23;12(8):1632. doi: 10.3390/biomedicines12081632. Biomedicines. 2024. PMID: 39200097 Free PMC article.
-
New understandings of the genetic regulatory relationship between non-coding RNAs and m6A modification.Front Genet. 2023 Dec 6;14:1270983. doi: 10.3389/fgene.2023.1270983. eCollection 2023. Front Genet. 2023. PMID: 38125749 Free PMC article. Review.
-
The Roles of RNA N6-Methyladenosine in Regulating Stem Cell Fate.Front Cell Dev Biol. 2021 Nov 5;9:765635. doi: 10.3389/fcell.2021.765635. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34805173 Free PMC article. Review.
-
Role of m6A modification in female infertility and reproductive system diseases.Int J Biol Sci. 2022 May 16;18(9):3592-3604. doi: 10.7150/ijbs.69771. eCollection 2022. Int J Biol Sci. 2022. PMID: 35813486 Free PMC article. Review.
References
-
- Balacco D.L., Soller M. The m6A writer: rise of a machine for growing tasks. Biochemistry. 2019;58:363–378. - PubMed
-
- Basciani S., De Luca G., Dolci S., Brama M., Arizzi M., Mariani S., Rosano G., Spera G., Gnessi L. Platelet-derived growth factor receptor β-subtype regulates proliferation and migration of gonocytes. Endocrinology. 2008;149:6226–6235. - PubMed
-
- Chen M., Zhang L.J., Cui X.H., Lin X.W., Li Y.Q., Wang Y.Q., Wang Y.B., Qin Y., Chen D.H., Han C.S. Wt1 directs the lineage specification of Sertoli and granulosa cells by repressing Sf1 expression. Development. 2017;144:44–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

