Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity
- PMID: 33051211
- PMCID: PMC7834058
- DOI: 10.1074/jbc.RA120.015138
Coronavirus infection and PARP expression dysregulate the NAD metabolome: An actionable component of innate immunity
Abstract
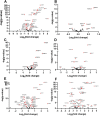
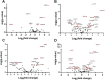
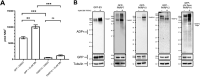
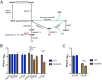
Poly(ADP-ribose) polymerase (PARP) superfamily members covalently link either a single ADP-ribose (ADPR) or a chain of ADPR units to proteins using NAD as the source of ADPR. Although the well-known poly(ADP-ribosylating) (PARylating) PARPs primarily function in the DNA damage response, many noncanonical mono(ADP-ribosylating) (MARylating) PARPs are associated with cellular antiviral responses. We recently demonstrated robust up-regulation of several PARPs following infection with murine hepatitis virus (MHV), a model coronavirus. Here we show that SARS-CoV-2 infection strikingly up-regulates MARylating PARPs and induces the expression of genes encoding enzymes for salvage NAD synthesis from nicotinamide (NAM) and nicotinamide riboside (NR), while down-regulating other NAD biosynthetic pathways. We show that overexpression of PARP10 is sufficient to depress cellular NAD and that the activities of the transcriptionally induced enzymes PARP7, PARP10, PARP12 and PARP14 are limited by cellular NAD and can be enhanced by pharmacological activation of NAD synthesis. We further demonstrate that infection with MHV induces a severe attack on host cell NAD+ and NADP+ Finally, we show that NAMPT activation, NAM, and NR dramatically decrease the replication of an MHV that is sensitive to PARP activity. These data suggest that the antiviral activities of noncanonical PARP isozyme activities are limited by the availability of NAD and that nutritional and pharmacological interventions to enhance NAD levels may boost innate immunity to coronaviruses.
Keywords: ADP-ribosylation; COVID-19; NAD biosynthesis; PARP; RNA-Seq; SARS-CoV-2; Severe acute respiratory syndrome coronavirus 2; gene transcription; interferon; nicotinamide adenine dinucleotide (NAD); plus-stranded RNA virus; poly(ADP-ribose) polymerase; post-translational modification (PTM); transcriptomics.
© 2020 Heer et al.
Conflict of interest statement
Conflict of interest—C. B. is chief scientific adviser of ChromaDex and owns shares of ChromaDex stock. C. B., S. A. J. T., S. P., and A. R. F. filed an invention disclosure on uses of NAD-boosting with respect to protection against coronavirus infection.
Figures

Update of
-
Coronavirus infection and PARP expression dysregulate the NAD Metabolome: an actionable component of innate immunity.bioRxiv [Preprint]. 2020 Oct 6:2020.04.17.047480. doi: 10.1101/2020.04.17.047480. bioRxiv. 2020. Update in: J Biol Chem. 2020 Dec 25;295(52):17986-17996. doi: 10.1074/jbc.RA120.015138. PMID: 32511303 Free PMC article. Updated. Preprint.
Similar articles
-
Coronavirus infection and PARP expression dysregulate the NAD Metabolome: an actionable component of innate immunity.bioRxiv [Preprint]. 2020 Oct 6:2020.04.17.047480. doi: 10.1101/2020.04.17.047480. bioRxiv. 2020. Update in: J Biol Chem. 2020 Dec 25;295(52):17986-17996. doi: 10.1074/jbc.RA120.015138. PMID: 32511303 Free PMC article. Updated. Preprint.
-
The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression.PLoS Pathog. 2019 May 16;15(5):e1007756. doi: 10.1371/journal.ppat.1007756. eCollection 2019 May. PLoS Pathog. 2019. PMID: 31095648 Free PMC article.
-
Mechanisms governing PARP expression, localization, and activity in cells.Crit Rev Biochem Mol Biol. 2020 Dec;55(6):541-554. doi: 10.1080/10409238.2020.1818686. Epub 2020 Sep 23. Crit Rev Biochem Mol Biol. 2020. PMID: 32962438
-
The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation.Viruses. 2020 Mar 31;12(4):384. doi: 10.3390/v12040384. Viruses. 2020. PMID: 32244383 Free PMC article. Review.
-
When PARPs Meet Antiviral Innate Immunity.Trends Microbiol. 2021 Sep;29(9):776-778. doi: 10.1016/j.tim.2021.01.002. Epub 2021 Jan 19. Trends Microbiol. 2021. PMID: 33483164 Review.
Cited by
-
Unique Mutations in the Murine Hepatitis Virus Macrodomain Differentially Attenuate Virus Replication, Indicating Multiple Roles for the Macrodomain in Coronavirus Replication.J Virol. 2021 Jul 12;95(15):e0076621. doi: 10.1128/JVI.00766-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011547 Free PMC article.
-
Delineating the SARS-CoV-2 Induced Interplay between the Host Immune System and the DNA Damage Response Network.Vaccines (Basel). 2022 Oct 20;10(10):1764. doi: 10.3390/vaccines10101764. Vaccines (Basel). 2022. PMID: 36298629 Free PMC article. Review.
-
Metabolic Signatures Associated with Severity in Hospitalized COVID-19 Patients.Int J Mol Sci. 2021 Apr 30;22(9):4794. doi: 10.3390/ijms22094794. Int J Mol Sci. 2021. PMID: 33946479 Free PMC article.
-
HTRF-based assay for detection of mono-ADP-ribosyl hydrolyzing macrodomains and inhibitor screening.iScience. 2024 Jun 20;27(7):110333. doi: 10.1016/j.isci.2024.110333. eCollection 2024 Jul 19. iScience. 2024. PMID: 39055912 Free PMC article.
-
The impact of aging and oxidative stress in metabolic and nervous system disorders: programmed cell death and molecular signal transduction crosstalk.Front Immunol. 2023 Nov 8;14:1273570. doi: 10.3389/fimmu.2023.1273570. eCollection 2023. Front Immunol. 2023. PMID: 38022638 Free PMC article. Review.
References
-
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sinica B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. 32292689. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

