Feedback of extended panel sequencing in 1530 patients referred for suspicion of hereditary predisposition to adult cancers
- PMID: 33047316
- PMCID: PMC7821123
- DOI: 10.1111/cge.13864
Feedback of extended panel sequencing in 1530 patients referred for suspicion of hereditary predisposition to adult cancers
Abstract
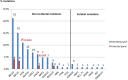
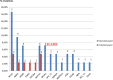
High-throughput sequencing analysis represented both a medical diagnosis and technological revolution. Gene panel analysis is now routinely performed in the exploration of hereditary predisposition to cancer, which is becoming increasingly heterogeneous, both clinically and molecularly. We present 1530 patients with suspicion of hereditary predisposition to cancer, for which two types of analyses were performed: a) oriented according to the clinical presentation (n = 417), or b) extended to genes involved in hereditary predisposition to adult cancer (n = 1113). Extended panel analysis had a higher detection rate compared to oriented analysis in hereditary predisposition to breast / ovarian cancer (P < .001) and in digestive cancers (P < .094) (respectively 15% vs 5% and 19.3%, vs 12.5%). This higher detection is explained by the inclusion of moderate penetrance genes, as well as the identification of incident mutations and double mutations. Our study underscores the utility of proposing extended gene panel analysis to patients with suspicion of hereditary predisposition to adult cancer.
Keywords: HBOC; HNPCC; double mutation; incidental findings ATM; panel sequencing; predisposition to cancer.
© 2020 The Authors. Clinical Genetics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare non conflict of interest. The study has been approved by personal protection committee (IRB: 2020 / CE57)
Figures

Similar articles
-
Gene-panel testing of breast and ovarian cancer patients identifies a recurrent RAD51C duplication.Clin Genet. 2018 Mar;93(3):595-602. doi: 10.1111/cge.13123. Epub 2018 Jan 12. Clin Genet. 2018. PMID: 28802053
-
Exon splicing analysis of intronic variants in multigene cancer panel testing for hereditary breast/ovarian cancer.Cancer Sci. 2020 Oct;111(10):3912-3925. doi: 10.1111/cas.14600. Epub 2020 Sep 2. Cancer Sci. 2020. PMID: 32761968 Free PMC article.
-
Identification of germline pathogenic variants in DNA damage repair genes by a next-generation sequencing multigene panel in BRCAX patients.Clin Biochem. 2020 Feb;76:17-23. doi: 10.1016/j.clinbiochem.2019.11.014. Epub 2019 Nov 28. Clin Biochem. 2020. PMID: 31786208
-
Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care.Breast Cancer Res Treat. 2016 Dec;160(3):393-410. doi: 10.1007/s10549-016-4003-9. Epub 2016 Oct 12. Breast Cancer Res Treat. 2016. PMID: 27734215 Review.
-
Current Approaches to Germline Cancer Genetic Testing.Annu Rev Med. 2020 Jan 27;71:85-102. doi: 10.1146/annurev-med-052318-101009. Epub 2019 Nov 22. Annu Rev Med. 2020. PMID: 31756137 Review.
Cited by
-
Atypical ATMs: Broadening the phenotypic spectrum of ATM-associated hereditary cancer.Front Oncol. 2023 Feb 14;13:1068110. doi: 10.3389/fonc.2023.1068110. eCollection 2023. Front Oncol. 2023. PMID: 36865800 Free PMC article.
-
Managing gastric cancer risk in lynch syndrome: controversies and recommendations.Fam Cancer. 2022 Jan;21(1):75-78. doi: 10.1007/s10689-021-00235-3. Epub 2021 Feb 21. Fam Cancer. 2022. PMID: 33611683 Free PMC article. Review. No abstract available.
-
Identification of a novel pathogenic variant in PALB2 and BARD1 genes by a multigene sequencing panel in triple negative breast cancer in Morocco.J Genomics. 2021 Sep 18;9:43-54. doi: 10.7150/jgen.61713. eCollection 2021. J Genomics. 2021. PMID: 34646395 Free PMC article.
-
Overview of the Genetic Causes of Hereditary Breast and Ovarian Cancer Syndrome in a Large French Patient Cohort.Cancers (Basel). 2023 Jun 29;15(13):3420. doi: 10.3390/cancers15133420. Cancers (Basel). 2023. PMID: 37444530 Free PMC article.
References
-
- Nielsen FC, van Overeem Hansen T, Sørensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat. Rev Cancer. 2016;16:599‐612. - PubMed
-
- Valle L. Recent discoveries in the genetics of familial colorectal cancer and polyposis. Clin. Gastroenterol. Hepatol. 2017;15:809‐819. - PubMed
-
- Sokolenko AP et al. Identification of novel hereditary cancer genes by whole exome sequencing. Cancer Lett. 2015;369:274‐288. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

