Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium
- PMID: 33046696
- PMCID: PMC7550582
- DOI: 10.1038/s41467-020-18781-2
Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium
Abstract
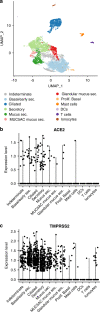
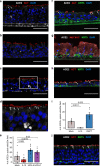
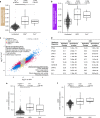
Coronavirus disease 2019 (COVID-19) is caused by SARS-CoV-2, an emerging virus that utilizes host proteins ACE2 and TMPRSS2 as entry factors. Understanding the factors affecting the pattern and levels of expression of these genes is important for deeper understanding of SARS-CoV-2 tropism and pathogenesis. Here we explore the role of genetics and co-expression networks in regulating these genes in the airway, through the analysis of nasal airway transcriptome data from 695 children. We identify expression quantitative trait loci for both ACE2 and TMPRSS2, that vary in frequency across world populations. We find TMPRSS2 is part of a mucus secretory network, highly upregulated by type 2 (T2) inflammation through the action of interleukin-13, and that the interferon response to respiratory viruses highly upregulates ACE2 expression. IL-13 and virus infection mediated effects on ACE2 expression were also observed at the protein level in the airway epithelium. Finally, we define airway responses to common coronavirus infections in children, finding that these infections generate host responses similar to other viral species, including upregulation of IL6 and ACE2. Our results reveal possible mechanisms influencing SARS-CoV-2 infectivity and COVID-19 clinical outcomes.
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Type 2 and interferon inflammation strongly regulate SARS-CoV-2 related gene expression in the airway epithelium.bioRxiv [Preprint]. 2020 Apr 10:2020.04.09.034454. doi: 10.1101/2020.04.09.034454. bioRxiv. 2020. Update in: Nat Commun. 2020 Oct 12;11(1):5139. doi: 10.1038/s41467-020-18781-2. PMID: 32511326 Free PMC article. Updated. Preprint.
Similar articles
-
Expression Pattern of the SARS-CoV-2 Entry Genes ACE2 and TMPRSS2 in the Respiratory Tract.Viruses. 2020 Oct 16;12(10):1174. doi: 10.3390/v12101174. Viruses. 2020. PMID: 33081421 Free PMC article.
-
Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue.Eur Respir J. 2020 Sep 3;56(3):2001123. doi: 10.1183/13993003.01123-2020. Print 2020 Sep. Eur Respir J. 2020. PMID: 32675206 Free PMC article.
-
SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues.Cell. 2020 May 28;181(5):1016-1035.e19. doi: 10.1016/j.cell.2020.04.035. Epub 2020 Apr 27. Cell. 2020. PMID: 32413319 Free PMC article.
-
Role of Genetic Variants and Gene Expression in the Susceptibility and Severity of COVID-19.Ann Lab Med. 2021 Mar 1;41(2):129-138. doi: 10.3343/alm.2021.41.2.129. Ann Lab Med. 2021. PMID: 33063674 Free PMC article. Review.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
Cited by
-
The type 2 asthma mediator IL-13 inhibits SARS-CoV-2 infection of bronchial epithelium.bioRxiv [Preprint]. 2021 Feb 25:2021.02.25.432762. doi: 10.1101/2021.02.25.432762. bioRxiv. 2021. Update in: Am J Respir Cell Mol Biol. 2022 Apr;66(4):391-401. doi: 10.1165/rcmb.2021-0364OC. PMID: 33655249 Free PMC article. Updated. Preprint.
-
COVID-19: From pathogenesis models to the first drug trials.Microb Biotechnol. 2020 Sep;13(5):1289-1299. doi: 10.1111/1751-7915.13611. Epub 2020 Jun 23. Microb Biotechnol. 2020. PMID: 32573950 Free PMC article. Review.
-
Immunosuppressants exert antiviral effects against influenza A(H1N1)pdm09 virus via inhibition of nucleic acid synthesis, mRNA splicing, and protein stability.Virulence. 2024 Dec;15(1):2301242. doi: 10.1080/21505594.2023.2301242. Epub 2024 Feb 6. Virulence. 2024. PMID: 38170681 Free PMC article.
-
Combination of Enrichment Using Gene Ontology and Transcriptomic Analysis Revealed Contribution of Interferon Signaling to Severity of COVID-19.Interdiscip Perspect Infect Dis. 2022 Apr 11;2022:3515001. doi: 10.1155/2022/3515001. eCollection 2022. Interdiscip Perspect Infect Dis. 2022. PMID: 35422859 Free PMC article.
-
Haplotype-aware modeling of cis-regulatory effects highlights the gaps remaining in eQTL data.Nat Commun. 2024 Jan 15;15(1):522. doi: 10.1038/s41467-024-44710-8. Nat Commun. 2024. PMID: 38225224 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 ES015794/ES/NIEHS NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- HL128439/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R01 HL141992/HL/NHLBI NIH HHS/United States
- UM1 HG008901/HG/NHGRI NIH HHS/United States
- R01 HL141845/HL/NHLBI NIH HHS/United States
- HL107202/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- U01 HG009080/HG/NHGRI NIH HHS/United States
- HL138626/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R01 HL117626/HL/NHLBI NIH HHS/United States
- HL135156/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- HL132821/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- P01 HL107202/HL/NHLBI NIH HHS/United States
- K01 HL140218/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- HL117004/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R01 HL135156/HL/NHLBI NIH HHS/United States
- T32 GM007546/GM/NIGMS NIH HHS/United States
- MD010443/U.S. Department of Health & Human Services | NIH | National Institute on Minority Health and Health Disparities (NIMHD)/International
- R01 HL128439/HL/NHLBI NIH HHS/United States
- R01 HL117004/HL/NHLBI NIH HHS/United States
- P60 MD006902/MD/NIMHD NIH HHS/United States
- HHSN268201600032C/HL/NHLBI NIH HHS/United States
- U24 HG008956/HG/NHGRI NIH HHS/United States
- P01 HL132821/HL/NHLBI NIH HHS/United States
- R01 MD010443/MD/NIMHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

