The E3 ubiquitin ligase HectD3 attenuates cardiac hypertrophy and inflammation in mice
- PMID: 33037313
- PMCID: PMC7547098
- DOI: 10.1038/s42003-020-01289-2
The E3 ubiquitin ligase HectD3 attenuates cardiac hypertrophy and inflammation in mice
Abstract
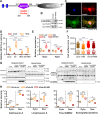
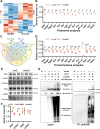
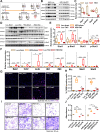
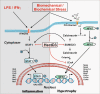
Myocardial inflammation has recently been recognized as a distinct feature of cardiac hypertrophy and heart failure. HectD3, a HECT domain containing E3 ubiquitin ligase has previously been investigated in the host defense against infections as well as neuroinflammation; its cardiac function however is still unknown. Here we show that HectD3 simultaneously attenuates Calcineurin-NFAT driven cardiomyocyte hypertrophy and the pro-inflammatory actions of LPS/interferon-γ via its cardiac substrates SUMO2 and Stat1, respectively. AAV9-mediated overexpression of HectD3 in mice in vivo not only reduced cardiac SUMO2/Stat1 levels and pathological hypertrophy but also largely abolished macrophage infiltration and fibrosis induced by pressure overload. Taken together, we describe a novel cardioprotective mechanism involving the ubiquitin ligase HectD3, which links anti-hypertrophic and anti-inflammatory effects via dual regulation of SUMO2 and Stat1. In a broader perspective, these findings support the notion that cardiomyocyte growth and inflammation are more intertwined than previously anticipated.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Sumoylation-independent activation of Calcineurin-NFAT-signaling via SUMO2 mediates cardiomyocyte hypertrophy.Sci Rep. 2016 Oct 21;6:35758. doi: 10.1038/srep35758. Sci Rep. 2016. PMID: 27767176 Free PMC article.
-
Targeting E3 Ubiquitin Ligase WWP1 Prevents Cardiac Hypertrophy Through Destabilizing DVL2 via Inhibition of K27-Linked Ubiquitination.Circulation. 2021 Aug 31;144(9):694-711. doi: 10.1161/CIRCULATIONAHA.121.054827. Epub 2021 Jun 18. Circulation. 2021. PMID: 34139860
-
HECT (Homologous to the E6-AP Carboxyl Terminus)-Type Ubiquitin E3 Ligase ITCH Attenuates Cardiac Hypertrophy by Suppressing the Wnt/β-Catenin Signaling Pathway.Hypertension. 2020 Dec;76(6):1868-1878. doi: 10.1161/HYPERTENSIONAHA.120.15487. Epub 2020 Nov 2. Hypertension. 2020. PMID: 33131309
-
The role of post-translational modifications in cardiac hypertrophy.J Cell Mol Med. 2019 Jun;23(6):3795-3807. doi: 10.1111/jcmm.14330. Epub 2019 Apr 4. J Cell Mol Med. 2019. PMID: 30950211 Free PMC article. Review.
-
The role of E3 ubiquitin ligase HECTD3 in cancer and beyond.Cell Mol Life Sci. 2020 Apr;77(8):1483-1495. doi: 10.1007/s00018-019-03339-3. Epub 2019 Oct 21. Cell Mol Life Sci. 2020. PMID: 31637449 Free PMC article. Review.
Cited by
-
Ubiquitination and deubiquitination in the regulation of N6-methyladenosine functional molecules.J Mol Med (Berl). 2024 Mar;102(3):337-351. doi: 10.1007/s00109-024-02417-9. Epub 2024 Jan 30. J Mol Med (Berl). 2024. PMID: 38289385 Review.
-
ARV-771 Acts as an Inducer of Cell Cycle Arrest and Apoptosis to Suppress Hepatocellular Carcinoma Progression.Front Pharmacol. 2022 May 4;13:858901. doi: 10.3389/fphar.2022.858901. eCollection 2022. Front Pharmacol. 2022. PMID: 35600879 Free PMC article.
-
E3 ligase HECTD3 promotes RNA virus replication and virus-induced inflammation via K33-linked polyubiquitination of PKR.Cell Death Dis. 2023 Jul 4;14(7):396. doi: 10.1038/s41419-023-05923-9. Cell Death Dis. 2023. PMID: 37402711 Free PMC article.
-
UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumor T cell response in prostate cancer.J Clin Invest. 2023 Feb 15;133(4):e158352. doi: 10.1172/JCI158352. J Clin Invest. 2023. PMID: 36626227 Free PMC article.
-
TRIM26 Induces Ferroptosis to Inhibit Hepatic Stellate Cell Activation and Mitigate Liver Fibrosis Through Mediating SLC7A11 Ubiquitination.Front Cell Dev Biol. 2021 Mar 25;9:644901. doi: 10.3389/fcell.2021.644901. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869196 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

