Proteomic Evaluation of the Comorbidity-Inflammation Paradigm in Heart Failure With Preserved Ejection Fraction: Results From the PROMIS-HFpEF Study
- PMID: 33034202
- PMCID: PMC7686082
- DOI: 10.1161/CIRCULATIONAHA.120.045810
Proteomic Evaluation of the Comorbidity-Inflammation Paradigm in Heart Failure With Preserved Ejection Fraction: Results From the PROMIS-HFpEF Study
Abstract
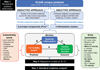
Background: A systemic proinflammatory state has been hypothesized to mediate the association between comorbidities and abnormal cardiac structure/function in heart failure with preserved ejection fraction (HFpEF). We conducted a proteomic analysis to investigate this paradigm.
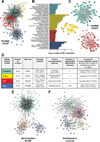
Methods: In 228 patients with HFpEF from the multicenter PROMIS-HFpEF study (Prevalence of Microvascular Dysfunction in Heart Failure With Preserved Ejection Fraction), 248 unique circulating proteins were quantified by a multiplex immunoassay (Olink) and used to recapitulate systemic inflammation. In a deductive approach, we performed principal component analysis to summarize 47 proteins known a priori to be involved in inflammation. In an inductive approach, we performed unbiased weighted coexpression network analyses of all 248 proteins to identify clusters of proteins that overrepresented inflammatory pathways. We defined comorbidity burden as the sum of 8 common HFpEF comorbidities. We used multivariable linear regression and statistical mediation analyses to determine whether and to what extent inflammation mediates the association of comorbidity burden with abnormal cardiac structure/function in HFpEF. We also externally validated our findings in an independent cohort of 117 HFpEF cases and 30 comorbidity controls without heart failure.
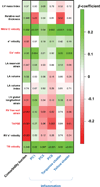
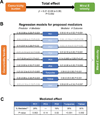
Results: Comorbidity burden was associated with abnormal cardiac structure/function and with principal components/clusters of inflammation proteins. Systemic inflammation was also associated with increased mitral E velocity, E/e' ratio, and tricuspid regurgitation velocity; and worse right ventricular function (tricuspid annular plane systolic excursion and right ventricular free wall strain). Inflammation mediated the association between comorbidity burden and mitral E velocity (proportion mediated 19%-35%), E/e' ratio (18%-29%), tricuspid regurgitation velocity (27%-41%), and tricuspid annular plane systolic excursion (13%) (P<0.05 for all), but not right ventricular free wall strain. TNFR1 (tumor necrosis factor receptor 1), UPAR (urokinase plasminogen activator receptor), IGFBP7 (insulin-like growth factor binding protein 7), and GDF-15 (growth differentiation factor-15) were the top individual proteins that mediated the relationship between comorbidity burden and echocardiographic parameters. In the validation cohort, inflammation was upregulated in HFpEF cases versus controls, and the most prominent inflammation protein cluster identified in PROMIS-HFpEF was also present in HFpEF cases (but not controls) in the validation cohort.
Conclusions: Proteins involved in inflammation form a conserved network in HFpEF across 2 independent cohorts and may mediate the association between comorbidity burden and echocardiographic indicators of worse hemodynamics and right ventricular dysfunction. These findings support the comorbidity-inflammation paradigm in HFpEF.
Keywords: analysis; biomarkers; comorbidity; echocardiography; heart failure; inflammation.
Figures
Similar articles
-
A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation.J Am Coll Cardiol. 2013 Jul 23;62(4):263-71. doi: 10.1016/j.jacc.2013.02.092. Epub 2013 May 15. J Am Coll Cardiol. 2013. PMID: 23684677 Review.
-
Impaired systolic function by strain imaging in heart failure with preserved ejection fraction.J Am Coll Cardiol. 2014 Feb 11;63(5):447-56. doi: 10.1016/j.jacc.2013.09.052. Epub 2013 Oct 30. J Am Coll Cardiol. 2014. PMID: 24184245 Free PMC article. Clinical Trial.
-
Right ventricular function in heart failure with preserved ejection fraction: a community-based study.Circulation. 2014 Dec 23;130(25):2310-20. doi: 10.1161/CIRCULATIONAHA.113.008461. Epub 2014 Nov 12. Circulation. 2014. PMID: 25391518 Free PMC article.
-
The impact of right ventricular dysfunction on the effectiveness of beta-blockers in heart failure with preserved ejection fraction.J Cardiol. 2020 Oct;76(4):325-334. doi: 10.1016/j.jjcc.2020.05.001. Epub 2020 May 28. J Cardiol. 2020. PMID: 32475652
-
Insight into the Pro-inflammatory and Profibrotic Role of Macrophage in Heart Failure With Preserved Ejection Fraction.J Cardiovasc Pharmacol. 2020 Sep;76(3):276-285. doi: 10.1097/FJC.0000000000000858. J Cardiovasc Pharmacol. 2020. PMID: 32501838 Review.
Cited by
-
Evaluating the adverse outcome of subtypes of heart failure with preserved ejection fraction defined by machine learning: A systematic review focused on defining high risk phenogroups.EXCLI J. 2022 Feb 22;21:487-518. doi: 10.17179/excli2021-4572. eCollection 2022. EXCLI J. 2022. PMID: 35391918 Free PMC article. Review.
-
Functional and Metabolic Imaging in Heart Failure with Preserved Ejection Fraction: Promises, Challenges, and Clinical Utility.Cardiovasc Drugs Ther. 2023 Apr;37(2):379-399. doi: 10.1007/s10557-022-07355-7. Epub 2022 Jul 26. Cardiovasc Drugs Ther. 2023. PMID: 35881280 Free PMC article. Review.
-
Biomarkers in Heart Failure with Preserved Ejection Fraction.Card Fail Rev. 2022 Jun 23;8:e20. doi: 10.15420/cfr.2021.37. eCollection 2022 Jan. Card Fail Rev. 2022. PMID: 35815256 Free PMC article. Review.
-
Obesity, Preserved Ejection Fraction Heart Failure, and Left Ventricular Remodeling.J Clin Med. 2023 May 8;12(9):3341. doi: 10.3390/jcm12093341. J Clin Med. 2023. PMID: 37176781 Free PMC article. Review.
-
From Systemic Inflammation to Myocardial Fibrosis: The Heart Failure With Preserved Ejection Fraction Paradigm Revisited.Circ Res. 2021 May 14;128(10):1451-1467. doi: 10.1161/CIRCRESAHA.121.318159. Epub 2021 May 13. Circ Res. 2021. PMID: 33983831 Free PMC article. Review.
References
-
- Paulus WJ, Tschope C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J Am Coll Cardiol. 2013;62:263–271. - PubMed
-
- Komajda M, Carson PE, Hetzel S, McKelvie R, McMurray J, Ptaszynska A, Zile MR, Demets D, Massie BM. Factors associated with outcome in heart failure with preserved ejection fraction: findings from the Irbesartan in Heart Failure with Preserved Ejection Fraction Study (I-PRESERVE). Circ Heart Fail. 2011;4:27–35. - PubMed
-
- Kristensen SL, Mogensen UM, Jhund PS, Petrie MC, Preiss D, Win S, Kober L, McKelvie RS, Zile MR, Anand IS, Komajda M, Gottdiener JS, Carson PE, McMurray JJ. Clinical and Echocardiographic Characteristics and Cardiovascular Outcomes According to Diabetes Status in Patients With Heart Failure and Preserved Ejection Fraction: A Report From the I-Preserve Trial (Irbesartan in Heart Failure With Preserved Ejection Fraction). Circulation. 2017;135:724–735. - PubMed
-
- Ather S, Chan W, Bozkurt B, Aguilar D, Ramasubbu K, Zachariah AA, Wehrens XH, Deswal A. Impact of noncardiac comorbidities on morbidity and mortality in a predominantly male population with heart failure and preserved versus reduced ejection fraction. J Am Coll Cardiol. 2012;59:998–1005. - PMC - PubMed
-
- Hage C, Michaelsson E, Linde C, Donal E, Daubert JC, Gan LM, Lund LH. Inflammatory Biomarkers Predict Heart Failure Severity and Prognosis in Patients With Heart Failure With Preserved Ejection Fraction: A Holistic Proteomic Approach. Circ Cardiovasc Genet. 2017;10:e001633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

