A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner
- PMID: 33025012
- PMCID: PMC8065239
- DOI: 10.1182/blood.2020006779
A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner
Erratum in
-
Elsaid R, Meunier S, Burlen-Defranoux O, et al. A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner. Blood. 2021;137(8):1024-1036.Blood. 2022 Sep 22;140(12):1451. doi: 10.1182/blood.2022017223. Blood. 2022. PMID: 36136354 Free PMC article. No abstract available.
Abstract
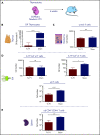
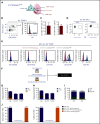
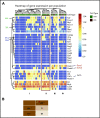
During embryonic development, multiple waves of hematopoietic progenitors with distinct lineage potential are differentially regulated in time and space. Two different waves of thymic progenitors colonize the fetal thymus where they contribute to thymic organogenesis and homeostasis. The origin, the lineage differentiation potential of the first wave, and their relative contribution in shaping the thymus architecture, remained, however, unclear. Here, we show that the first wave of thymic progenitors comprises a unique population of bipotent T and innatel lymphoid cells (T/ILC), generating a lymphoid tissue inducer cells (LTi's), in addition to invariant Vγ5+ T cells. Transcriptional analysis revealed that innate lymphoid gene signatures and, more precisely, the LTi-associated transcripts were expressed in the first, but not in the second, wave of thymic progenitors. Depletion of early thymic progenitors in a temporally controlled manner showed that the progeny of the first wave is indispensable for the differentiation of autoimmune regulator-expressing medullary thymic epithelial cells (mTECs). We further show that these progenitors are of strict hematopoietic stem cell origin, despite the overlap between lymphopoiesis initiation and the transient expression of lymphoid-associated transcripts in yolk sac (YS) erythromyeloid-restricted precursors. Our work highlights the relevance of the developmental timing on the emergence of different lymphoid subsets, required for the establishment of a functionally diverse immune system.
© 2021 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



Comment in
-
Layered immunity of the developing thymus.Blood. 2021 Feb 25;137(8):1003-1004. doi: 10.1182/blood.2020009207. Blood. 2021. PMID: 33630049 Free PMC article.
Similar articles
-
Clonal analysis reveals uniformity in the molecular profile and lineage potential of CCR9(+) and CCR9(-) thymus-settling progenitors.J Immunol. 2011 May 1;186(9):5227-35. doi: 10.4049/jimmunol.1002686. Epub 2011 Mar 18. J Immunol. 2011. PMID: 21421850 Free PMC article.
-
Single-Cell RNA Sequencing Resolves Spatiotemporal Development of Pre-thymic Lymphoid Progenitors and Thymus Organogenesis in Human Embryos.Immunity. 2019 Nov 19;51(5):930-948.e6. doi: 10.1016/j.immuni.2019.09.008. Epub 2019 Oct 8. Immunity. 2019. PMID: 31604687
-
Differential requirement for CCR4 in the maintenance but not establishment of the invariant Vγ5(+) dendritic epidermal T-cell pool.PLoS One. 2013 Sep 12;8(9):e74019. doi: 10.1371/journal.pone.0074019. eCollection 2013. PLoS One. 2013. PMID: 24069263 Free PMC article.
-
New Molecular Insights into Immune Cell Development.Annu Rev Immunol. 2019 Apr 26;37:497-519. doi: 10.1146/annurev-immunol-042718-041319. Annu Rev Immunol. 2019. PMID: 31026413 Review.
-
Adult thymic epithelial cell (TEC) progenitors and TEC stem cells: Models and mechanisms for TEC development and maintenance.Eur J Immunol. 2015 Nov;45(11):2985-93. doi: 10.1002/eji.201545844. Epub 2015 Sep 30. Eur J Immunol. 2015. PMID: 26362014 Review.
Cited by
-
Shaping immunity for life: Layered development of CD8+ T cells.Immunol Rev. 2023 May;315(1):108-125. doi: 10.1111/imr.13185. Epub 2023 Jan 18. Immunol Rev. 2023. PMID: 36653953 Free PMC article. Review.
-
Type 1 immunity enables neonatal thymic ILC1 production.Sci Adv. 2024 Jan 19;10(3):eadh5520. doi: 10.1126/sciadv.adh5520. Epub 2024 Jan 17. Sci Adv. 2024. PMID: 38232171 Free PMC article.
-
Lymphoid Tissue inducer (LTi) cell ontogeny and functioning in embryo and adult.Biomed J. 2021 Apr;44(2):123-132. doi: 10.1016/j.bj.2020.12.003. Epub 2020 Dec 10. Biomed J. 2021. PMID: 33849806 Free PMC article. Review.
-
Definitive hematopoiesis is dispensable to sustain erythrocytes and macrophages during zebrafish ontogeny.iScience. 2024 Jan 17;27(2):108922. doi: 10.1016/j.isci.2024.108922. eCollection 2024 Feb 16. iScience. 2024. PMID: 38327794 Free PMC article.
-
Hematopoietic Stem Cell Development in Mammalian Embryos.Adv Exp Med Biol. 2023;1442:1-16. doi: 10.1007/978-981-99-7471-9_1. Adv Exp Med Biol. 2023. PMID: 38228955 Review.
References
-
- Cumano A, Berthault C, Ramond C, et al. . New molecular insights into immune cell development. Annu Rev Immunol. 2019;37:497-519. - PubMed
-
- Berthault C, Ramond C, Burlen-Defranoux O, et al. . Asynchronous lineage priming determines commitment to T cell and B cell lineages in fetal liver. Nat Immunol. 2017;18(10):1139-1149. - PubMed
-
- Ramond C, Berthault C, Burlen-Defranoux O, et al. . Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol. 2014;15(1):27-35. - PubMed
-
- Bhandoola A, von Boehmer H, Petrie HT, Zúñiga-Pflücker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26(6):678-689. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

