Network-based identification genetic effect of SARS-CoV-2 infections to Idiopathic pulmonary fibrosis (IPF) patients
- PMID: 33024988
- PMCID: PMC7665362
- DOI: 10.1093/bib/bbaa235
Network-based identification genetic effect of SARS-CoV-2 infections to Idiopathic pulmonary fibrosis (IPF) patients
Abstract
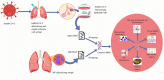
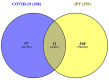
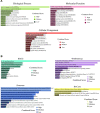
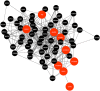
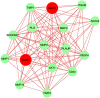
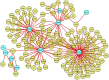
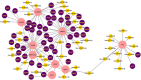
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is accountable for the cause of coronavirus disease (COVID-19) that causes a major threat to humanity. As the spread of the virus is probably getting out of control on every day, the epidemic is now crossing the most dreadful phase. Idiopathic pulmonary fibrosis (IPF) is a risk factor for COVID-19 as patients with long-term lung injuries are more likely to suffer in the severity of the infection. Transcriptomic analyses of SARS-CoV-2 infection and IPF patients in lung epithelium cell datasets were selected to identify the synergistic effect of SARS-CoV-2 to IPF patients. Common genes were identified to find shared pathways and drug targets for IPF patients with COVID-19 infections. Using several enterprising Bioinformatics tools, protein-protein interactions (PPIs) network was designed. Hub genes and essential modules were detected based on the PPIs network. TF-genes and miRNA interaction with common differentially expressed genes and the activity of TFs are also identified. Functional analysis was performed using gene ontology terms and Kyoto Encyclopedia of Genes and Genomes pathway and found some shared associations that may cause the increased mortality of IPF patients for the SARS-CoV-2 infections. Drug molecules for the IPF were also suggested for the SARS-CoV-2 infections.
Keywords: SARS-CoV-2; differentially expressed genes; drug molecule; gene ontology; hub gene; idiopathic pulmonary fibrosis; protein–protein interactions.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients.Brief Bioinform. 2021 Sep 2;22(5):bbab115. doi: 10.1093/bib/bbab115. Brief Bioinform. 2021. PMID: 33847347 Free PMC article.
-
Bioinformatics-based investigation on the genetic influence between SARS-CoV-2 infections and idiopathic pulmonary fibrosis (IPF) diseases, and drug repurposing.Sci Rep. 2023 Mar 22;13(1):4685. doi: 10.1038/s41598-023-31276-6. Sci Rep. 2023. PMID: 36949176 Free PMC article.
-
MLBioIGE: integration and interplay of machine learning and bioinformatics approach to identify the genetic effect of SARS-COV-2 on idiopathic pulmonary fibrosis patients.Biol Methods Protoc. 2022 May 30;7(1):bpac013. doi: 10.1093/biomethods/bpac013. eCollection 2022. Biol Methods Protoc. 2022. PMID: 35734766 Free PMC article.
-
Identification of the effects of COVID-19 on patients with pulmonary fibrosis and lung cancer: a bioinformatics analysis and literature review.Sci Rep. 2022 Sep 26;12(1):16040. doi: 10.1038/s41598-022-20040-x. Sci Rep. 2022. PMID: 36163484 Free PMC article. Review.
-
Characteristics and outcomes of COVID-19 patients with IPF: A multi-center retrospective study.Respir Med Res. 2022 May;81:100900. doi: 10.1016/j.resmer.2022.100900. Epub 2022 Mar 3. Respir Med Res. 2022. PMID: 35338917 Free PMC article. Review.
Cited by
-
Differential gene expression profiling reveals potential biomarkers and pharmacological compounds against SARS-CoV-2: Insights from machine learning and bioinformatics approaches.Front Immunol. 2022 Aug 17;13:918692. doi: 10.3389/fimmu.2022.918692. eCollection 2022. Front Immunol. 2022. PMID: 36059456 Free PMC article.
-
Identifying the Effect of COVID-19 Infection in Multiple Myeloma and Diffuse Large B-Cell Lymphoma Patients Using Bioinformatics and System Biology.Comput Math Methods Med. 2022 Nov 23;2022:7017317. doi: 10.1155/2022/7017317. eCollection 2022. Comput Math Methods Med. 2022. PMID: 36466549 Free PMC article.
-
New Insight Into Mechanisms of Hepatic Encephalopathy: An Integrative Analysis Approach to Identify Molecular Markers and Therapeutic Targets.Bioinform Biol Insights. 2023 Feb 17;17:11779322231155068. doi: 10.1177/11779322231155068. eCollection 2023. Bioinform Biol Insights. 2023. PMID: 36814683 Free PMC article.
-
Single-cell RNA sequencing and multiple bioinformatics methods to identify the immunity and ferroptosis-related biomarkers of SARS-CoV-2 infections to ischemic stroke.Aging (Albany NY). 2023 Aug 21;15(16):8237-8257. doi: 10.18632/aging.204966. Epub 2023 Aug 21. Aging (Albany NY). 2023. PMID: 37606960 Free PMC article.
-
Identification of Key Genes Related to Immune Cells in Patients with COVID-19 Via Integrated Bioinformatics-Based Analysis.Biochem Genet. 2023 Dec;61(6):2650-2671. doi: 10.1007/s10528-023-10400-1. Epub 2023 May 24. Biochem Genet. 2023. PMID: 37222960 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

