Exploiting Replication Stress as a Novel Therapeutic Intervention
- PMID: 33020173
- PMCID: PMC7864862
- DOI: 10.1158/1541-7786.MCR-20-0651
Exploiting Replication Stress as a Novel Therapeutic Intervention
Abstract
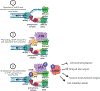
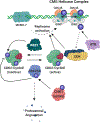
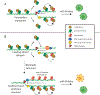
Ewing sarcoma is an aggressive pediatric tumor of the bone and soft tissue. The current standard of care is radiation and chemotherapy, and patients generally lack targeted therapies. One of the defining molecular features of this tumor type is the presence of significantly elevated levels of replication stress as compared with both normal cells and many other types of cancers, but the source of this stress is poorly understood. Tumors that harbor elevated levels of replication stress rely on the replication stress and DNA damage response pathways to retain viability. Understanding the source of the replication stress in Ewing sarcoma may reveal novel therapeutic targets. Ewing sarcomagenesis is complex, and in this review, we discuss the current state of our knowledge regarding elevated replication stress and the DNA damage response in Ewing sarcoma, one contributor to the disease process. We will also describe how these pathways are being successfully targeted therapeutically in other tumor types, and discuss possible novel, evidence-based therapeutic interventions in Ewing sarcoma. We hope that this consolidation will spark investigations that uncover new therapeutic targets and lead to the development of better treatment options for patients with Ewing sarcoma. IMPLICATIONS: This review uncovers new therapeutic targets in Ewing sarcoma and highlights replication stress as an exploitable vulnerability across multiple cancers.
©2020 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
Similar articles
-
Sequencing Overview of Ewing Sarcoma: A Journey across Genomic, Epigenomic and Transcriptomic Landscapes.Int J Mol Sci. 2015 Jul 16;16(7):16176-215. doi: 10.3390/ijms160716176. Int J Mol Sci. 2015. PMID: 26193259 Free PMC article. Review.
-
High-throughput RNAi screen in Ewing sarcoma cells identifies leucine rich repeats and WD repeat domain containing 1 (LRWD1) as a regulator of EWS-FLI1 driven cell viability.Gene. 2017 Jan 5;596:137-146. doi: 10.1016/j.gene.2016.10.021. Epub 2016 Oct 17. Gene. 2017. PMID: 27760381
-
The second European interdisciplinary Ewing sarcoma research summit--A joint effort to deconstructing the multiple layers of a complex disease.Oncotarget. 2016 Feb 23;7(8):8613-24. doi: 10.18632/oncotarget.6937. Oncotarget. 2016. PMID: 26802024 Free PMC article. Review.
-
[Functional genomics of Ewing sarcoma].Pathologe. 2017 Nov;38(Suppl 2):198-201. doi: 10.1007/s00292-017-0332-7. Pathologe. 2017. PMID: 28849372 Review. German.
-
Emerging therapies in Ewing sarcoma.Curr Opin Oncol. 2024 Jul 1;36(4):297-304. doi: 10.1097/CCO.0000000000001048. Epub 2024 May 22. Curr Opin Oncol. 2024. PMID: 38775200 Free PMC article. Review.
Cited by
-
WEE1 inhibition augments CDC7 (DDK) inhibitor-induced cell death in Ewing sarcoma by forcing premature mitotic entry and mitotic catastrophe.Cancer Res Commun. 2022 Jun;2(6):471-482. doi: 10.1158/2767-9764.crc-22-0130. Epub 2022 Jun 20. Cancer Res Commun. 2022. PMID: 36338546 Free PMC article.
-
Synergistic anticancer activity of combined ATR and ribonucleotide reductase inhibition in Ewing's sarcoma cells.J Cancer Res Clin Oncol. 2023 Sep;149(11):8605-8617. doi: 10.1007/s00432-023-04804-0. Epub 2023 Apr 25. J Cancer Res Clin Oncol. 2023. PMID: 37097390 Free PMC article.
-
Molecularly Defined Subsets of Ewing Sarcoma Tumors Differ in Their Responses to IGF1R and WEE1 Inhibition.Clin Cancer Res. 2023 Jan 17;29(2):458-471. doi: 10.1158/1078-0432.CCR-22-2587. Clin Cancer Res. 2023. PMID: 36394520 Free PMC article.
-
Cooperative treatment effectiveness of ATR and HSP90 inhibition in Ewing's sarcoma cells.Cell Biosci. 2021 Mar 20;11(1):57. doi: 10.1186/s13578-021-00571-y. Cell Biosci. 2021. PMID: 33743824 Free PMC article.
-
Upregulation of the Mevalonate Pathway through EWSR1-FLI1/EGR2 Regulatory Axis Confers Ewing Cells Exquisite Sensitivity to Statins.Cancers (Basel). 2022 May 8;14(9):2327. doi: 10.3390/cancers14092327. Cancers (Basel). 2022. PMID: 35565457 Free PMC article.
References
-
- Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2015;33(27):3036–46 doi 10.1200/jco.2014.59.5256. - DOI - PubMed
-
- Horowitz M, Delaney T, Malawer M, Woo S, Hicks M. Ewing’s sarcoma of bone and soft tissue and peripheral primitive neuroectodermal tumor Principles and practice of pediatric oncology. Philadelphia: Lippincott; 1997.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

