Molecular characterization of canine coronaviruses: an enteric and pantropic approach
- PMID: 33005986
- PMCID: PMC7529357
- DOI: 10.1007/s00705-020-04826-w
Molecular characterization of canine coronaviruses: an enteric and pantropic approach
Abstract
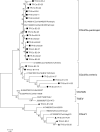
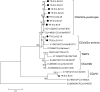
Canine coronavirus (CCoV) generally causes an infection with high morbidity and low mortality in dogs. In recent years, studies on coronaviruses have gained a momentum due to coronavirus outbreaks. Mutations in coronaviruses can result in deadly diseases in new hosts (such as SARS-CoV-2) or cause changes in organ-tissue affinity, as occurred with feline infectious peritonitis virus, exacerbating their pathogenesis. In recent studies on different types of CCoV, the pantropic strains characterized by hypervirulent and multi-systemic infections are believed to be emerging, in contrast to classical enteric coronavirus infections. In this study, we investigated emerging hypervirulent and multi-systemic CCoV strains using molecular and bioinformatic analysis, and examined differences between enteric and pantropic CCoV strains at the phylogenetic level. RT-PCR was performed with specific primers to identify the coronavirus M (membrane) and S (spike) genes, and samples were then subjected to DNA sequencing. In phylogenetic analysis, four out of 26 samples were classified as CCoV-1. The remaining 22 samples were all classified as CCoV-2a. In the CCoV-2a group, six samples were in branches close to enteric strains, and 16 samples were in the branches close to pantropic strains. Enteric and pantropic strains were compared by molecular genotyping of CCoV in dogs. Phylogenetic analysis of hypervirulent pantropic strains was carried out at the amino acid and nucleotide sequence levels. CCoV was found to be divergent from the original strain. This implies that some CCoV strains have become pantropic strains that cause multisystemic infections, and they should not be ruled out as the cause of severe diarrhea and multisystemic infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Characterization of pantropic canine coronavirus from Brazil.Vet J. 2014 Dec;202(3):659-62. doi: 10.1016/j.tvjl.2014.09.006. Epub 2014 Sep 16. Vet J. 2014. PMID: 25294661 Free PMC article.
-
Molecular characterization of a canine coronavirus NA/09 strain detected in a dog's organs.Arch Virol. 2012 Jan;157(1):171-5. doi: 10.1007/s00705-011-1141-6. Epub 2011 Oct 16. Arch Virol. 2012. PMID: 22002680 Free PMC article.
-
Infection of cats with atypical feline coronaviruses harbouring a truncated form of the canine type I non-structural ORF3 gene.Infect Genet Evol. 2013 Dec;20:488-94. doi: 10.1016/j.meegid.2013.09.024. Epub 2013 Oct 9. Infect Genet Evol. 2013. PMID: 24121017 Free PMC article.
-
Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins.Viruses. 2014 Aug 22;6(8):3363-76. doi: 10.3390/v6083363. Viruses. 2014. PMID: 25153347 Free PMC article. Review.
-
Genetic evolution of canine coronavirus and recent advances in prophylaxis.Vet Res. 2006 Mar-Apr;37(2):191-200. doi: 10.1051/vetres:2005053. Vet Res. 2006. PMID: 16472519 Review.
Cited by
-
Canine Coronavirus Activates Aryl Hydrocarbon Receptor during In Vitro Infection.Viruses. 2022 Nov 3;14(11):2437. doi: 10.3390/v14112437. Viruses. 2022. PMID: 36366535 Free PMC article.
-
Identification and phylogenetic analysis of two canine coronavirus strains.Anim Dis. 2021;1(1):10. doi: 10.1186/s44149-021-00013-9. Epub 2021 Jul 19. Anim Dis. 2021. PMID: 34778880 Free PMC article.
-
Effectiveness of the Fungal Metabolite 3-O-Methylfunicone towards Canine Coronavirus in a Canine Fibrosarcoma Cell Line (A72).Antibiotics (Basel). 2022 Nov 11;11(11):1594. doi: 10.3390/antibiotics11111594. Antibiotics (Basel). 2022. PMID: 36421238 Free PMC article.
-
All Microbiological Aspects of SARS-CoV-2 Virus.Eurasian J Med. 2022 Dec;54(Suppl1):106-114. doi: 10.5152/eurasianjmed.2022.22315. Eurasian J Med. 2022. PMID: 36655453 Free PMC article.
-
Detection and Molecular Characterization of Kobuviruses: An Agent of Canine Viral Diarrhea.Curr Microbiol. 2024 Aug 16;81(10):309. doi: 10.1007/s00284-024-03831-5. Curr Microbiol. 2024. PMID: 39150576
References
-
- Yesilbag K, Aytogu G. Coronavirus host divergence and novel coronavirus (Sars-CoV-2) outbreak. Clin Exp Ocul Trauma Infect. 2020;2(1):139–147.
-
- Enjuanes L, Brian D, Cavanagh D, et al. et al. Family Coronaviridae. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, et al.et al., editors. Virus taxonomy, classification and nomenclature of viruses. New York: Academic Press; 2000. pp. 835–849.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

