Studying the surfaces of bacteria using neutron scattering: finding new openings for antibiotics
- PMID: 33005925
- PMCID: PMC7609035
- DOI: 10.1042/BST20200320
Studying the surfaces of bacteria using neutron scattering: finding new openings for antibiotics
Abstract
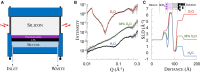
The use of neutrons as a scattering probe to investigate biological membranes has steadily grown in the past three decades, shedding light on the structure and behaviour of this ubiquitous and fundamental biological barrier. Meanwhile, the rise of antibiotic resistance has catalysed a renewed interest in understanding the mechanisms underlying the dynamics of antibiotics interaction with the bacterial cell envelope. It is widely recognised that the key reason behind the remarkable success of Gram-negative pathogens in developing antibiotic resistance lies in the effectiveness of their outer membrane (OM) in defending the cell from antibacterial compounds. Critical to its function, the highly asymmetric lipid distribution between the inner and outer bilayer leaflets of the OM, adds an extra level of complexity to the study of this crucial defence barrier. Here we review the opportunities offered by neutron scattering techniques, in particular reflectometry, to provide structural information on the interactions of antimicrobials with in vitro models of the OM. The differential sensitivity of neutrons towards hydrogen and deuterium makes them a unique probe to study the structure and behaviour of asymmetric membranes. Molecular-level understanding of the interactions between antimicrobials and the Gram-negative OM provides valuable insights that can aid drug development and broaden our knowledge of this critically important biological barrier.
Keywords: antibiotics; biological models; gram-negative bacteria; lipopolysaccharides; neutron scattering; outer membrane.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


Similar articles
-
Liquid crystalline bacterial outer membranes are critical for antibiotic susceptibility.Proc Natl Acad Sci U S A. 2018 Aug 7;115(32):E7587-E7594. doi: 10.1073/pnas.1803975115. Epub 2018 Jul 23. Proc Natl Acad Sci U S A. 2018. PMID: 30037998 Free PMC article.
-
Asymmetric phospholipid: lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic.J R Soc Interface. 2013 Oct 16;10(89):20130810. doi: 10.1098/rsif.2013.0810. Print 2013 Dec 6. J R Soc Interface. 2013. PMID: 24132206 Free PMC article.
-
Deuterium Labeling Strategies for Creating Contrast in Structure-Function Studies of Model Bacterial Outer Membranes Using Neutron Reflectometry.Methods Enzymol. 2016;566:231-52. doi: 10.1016/bs.mie.2015.05.020. Epub 2015 Jun 18. Methods Enzymol. 2016. PMID: 26791981
-
Lipopolysaccharides at Solid and Liquid Interfaces: Models for Biophysical Studies of the Gram-negative Bacterial Outer Membrane.Adv Colloid Interface Sci. 2022 Mar;301:102603. doi: 10.1016/j.cis.2022.102603. Epub 2022 Jan 23. Adv Colloid Interface Sci. 2022. PMID: 35093846 Review.
-
Outer Membrane Lipid Secretion and the Innate Immune Response to Gram-Negative Bacteria.Infect Immun. 2020 Jun 22;88(7):e00920-19. doi: 10.1128/IAI.00920-19. Print 2020 Jun 22. Infect Immun. 2020. PMID: 32253250 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

