The SAGA chromatin-modifying complex: the sum of its parts is greater than the whole
- PMID: 33004486
- PMCID: PMC7528701
- DOI: 10.1101/gad.341156.120
The SAGA chromatin-modifying complex: the sum of its parts is greater than the whole
Abstract
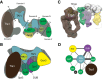
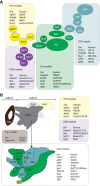
There are many large protein complexes involved in transcription in a chromatin context. However, recent studies on the SAGA coactivator complex are generating new paradigms for how the components of these complexes function, both independently and in concert. This review highlights the initial discovery of the canonical SAGA complex 23 years ago, our evolving understanding of its modular structure and the relevance of its modular nature for its coactivator function in gene regulation.
Keywords: DUB; HAT; SAGA; activators; adaptor complex; adaptors; chromatin; coactivator; structure; transcription.
© 2020 Soffers and Workman; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Structure of the transcription coactivator SAGA.Nature. 2020 Jan;577(7792):717-720. doi: 10.1038/s41586-020-1933-5. Epub 2020 Jan 22. Nature. 2020. PMID: 31969703 Free PMC article.
-
Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes.Mol Cell Biol. 2011 Feb;31(4):818-31. doi: 10.1128/MCB.00687-10. Epub 2010 Dec 13. Mol Cell Biol. 2011. PMID: 21149579 Free PMC article.
-
Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex.EMBO J. 2014 Nov 3;33(21):2534-46. doi: 10.15252/embj.201488638. Epub 2014 Sep 12. EMBO J. 2014. PMID: 25216679 Free PMC article.
-
SAGA and TFIID: Friends of TBP drifting apart.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194604. doi: 10.1016/j.bbagrm.2020.194604. Epub 2020 Jul 14. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32673655 Review.
-
Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes.Transcription. 2019 Feb;10(1):37-43. doi: 10.1080/21541264.2018.1530936. Epub 2018 Oct 30. Transcription. 2019. PMID: 30375921 Free PMC article. Review.
Cited by
-
Functional networks of the human bromodomain-containing proteins.Front Bioinform. 2022 Aug 10;2:835892. doi: 10.3389/fbinf.2022.835892. eCollection 2022. Front Bioinform. 2022. PMID: 36304339 Free PMC article.
-
UPS writes a new saga of SAGA.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194981. doi: 10.1016/j.bbagrm.2023.194981. Epub 2023 Aug 30. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37657588 Free PMC article. Review.
-
Structures and implications of TBP-nucleosome complexes.Proc Natl Acad Sci U S A. 2021 Jul 27;118(30):e2108859118. doi: 10.1073/pnas.2108859118. Proc Natl Acad Sci U S A. 2021. PMID: 34301908 Free PMC article.
-
Increased Agrobacterium-mediated transformation of Saccharomyces cerevisiae after deletion of the yeast ADA2 gene.Lett Appl Microbiol. 2022 Feb;74(2):228-237. doi: 10.1111/lam.13605. Epub 2021 Nov 27. Lett Appl Microbiol. 2022. PMID: 34816457 Free PMC article.
-
The SAGA complex regulates early steps in transcription via its deubiquitylase module subunit USP22.EMBO J. 2021 Aug 16;40(16):e102509. doi: 10.15252/embj.2019102509. Epub 2021 Jun 22. EMBO J. 2021. PMID: 34155658 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
