Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement
- PMID: 33003317
- PMCID: PMC7582545
- DOI: 10.3390/ijms21197180
Systematic Analysis of Gibberellin Pathway Components in Medicago truncatula Reveals the Potential Application of Gibberellin in Biomass Improvement
Abstract
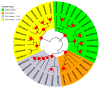
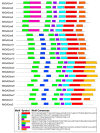
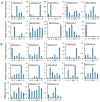
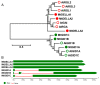
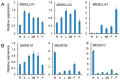
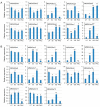
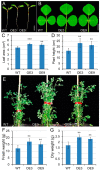
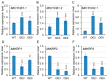
Gibberellins (GAs), a class of phytohormones, act as an essential natural regulator of plant growth and development. Many studies have shown that GA is related to rhizobial infection and nodule organogenesis in legume species. However, thus far, GA metabolism and signaling components are largely unknown in the model legume Medicago truncatula. In this study, a genome-wide analysis of GA metabolism and signaling genes was carried out. In total 29 components, including 8 MtGA20ox genes, 2 MtGA3ox genes, 13 MtGA2ox genes, 3 MtGID1 genes, and 3 MtDELLA genes were identified in M. truncatula genome. Expression profiles revealed that most members of MtGAox, MtGID1, and MtDELLA showed tissue-specific expression patterns. In addition, the GA biosynthesis and deactivation genes displayed a feedback regulation on GA treatment, respectively. Yeast two-hybrid assays showed that all the three MtGID1s interacted with MtDELLA1 and MtDELLA2, suggesting that the MtGID1s are functional GA receptors. More importantly, M. truncatula exhibited increased plant height and biomass by ectopic expression of the MtGA20ox1, suggesting that enhanced GA response has the potential for forage improvement.
Keywords: Medicago truncatula; expression analysis; forage improvement; gibberellins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Gibberellin dynamics governing nodulation revealed using GIBBERELLIN PERCEPTION SENSOR 2 in Medicago truncatula lateral organs.Plant Cell. 2024 Oct 3;36(10):4442-4456. doi: 10.1093/plcell/koae201. Plant Cell. 2024. PMID: 39012965 Free PMC article.
-
MtGA2ox10 encoding C20-GA2-oxidase regulates rhizobial infection and nodule development in Medicago truncatula.Sci Rep. 2019 Apr 11;9(1):5952. doi: 10.1038/s41598-019-42407-3. Sci Rep. 2019. PMID: 30976084 Free PMC article.
-
Cytokinins and the CRE1 receptor influence endogenous gibberellin levels in Medicago truncatula.Plant Signal Behav. 2018 Feb 1;13(2):e1428513. doi: 10.1080/15592324.2018.1428513. Epub 2018 Feb 6. Plant Signal Behav. 2018. PMID: 29373072 Free PMC article.
-
Root Development in Medicago truncatula: Lessons from Genetics to Functional Genomics.Methods Mol Biol. 2018;1822:205-239. doi: 10.1007/978-1-4939-8633-0_15. Methods Mol Biol. 2018. PMID: 30043307 Review.
-
Gibberellin signaling in plants.Development. 2013 Mar;140(6):1147-51. doi: 10.1242/dev.087650. Development. 2013. PMID: 23444347 Review.
Cited by
-
Salt Stress Enhances Early Symbiotic Gene Expression in Medicago truncatula and Induces a Stress-Specific Set of Rhizobium-Responsive Genes.Mol Plant Microbe Interact. 2021 Aug;34(8):904-921. doi: 10.1094/MPMI-01-21-0019-R. Epub 2021 Sep 8. Mol Plant Microbe Interact. 2021. PMID: 33819071 Free PMC article.
-
Identification of loci controlling timing of stem elongation in red clover using genotyping by sequencing of pooled phenotypic extremes.Mol Genet Genomics. 2022 Nov;297(6):1587-1600. doi: 10.1007/s00438-022-01942-x. Epub 2022 Aug 24. Mol Genet Genomics. 2022. PMID: 36001174 Free PMC article.
-
Gibberellin Oxidase Gene Family in L. chinense: Genome-Wide Identification and Gene Expression Analysis.Int J Mol Sci. 2021 Jul 2;22(13):7167. doi: 10.3390/ijms22137167. Int J Mol Sci. 2021. PMID: 34281216 Free PMC article.
-
The GA 20-Oxidase Encoding Gene MSD1 Controls the Main Stem Elongation in Medicago truncatula.Front Plant Sci. 2021 Aug 4;12:709625. doi: 10.3389/fpls.2021.709625. eCollection 2021. Front Plant Sci. 2021. PMID: 34421956 Free PMC article.
-
Plant Development and Crop Yield: The Role of Gibberellins.Plants (Basel). 2022 Oct 9;11(19):2650. doi: 10.3390/plants11192650. Plants (Basel). 2022. PMID: 36235516 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

